Gene
KWMTBOMO15592 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010723
Annotation
small_nuclear_ribonucleoprotein_sm_d2_[Bombyx_mori]
Full name
Small nuclear ribonucleoprotein Sm D2
+ More
Probable small nuclear ribonucleoprotein Sm D2
Probable small nuclear ribonucleoprotein Sm D2
Alternative Name
snRNP core protein D2
Location in the cell
Nuclear Reliability : 2.796
Sequence
CDS
ATGACCCTAGAAGAGCTATCTAAAATCGAAGAGGAAGAGTTTAGTACCGGACCTCTTTCCGTGCTCACGCAGTCAGTAAAGAACAACACACAAGTTCTGATAAATTGCCGTAACAATAAGAAACTCCTGGGTAGAGTGAAGGCTTTCGACCGGCATTGTAACATGGTATTGGAGAATGTTAAGGAAATGTGGACTGAGGTACCGAGAACTGGCAAAGGAAAGAAGGGCAAAGCTGTAAACAAGGACAAATTCATCTCCAAAATGTTCCTCCGCGGTGATTCTGTGATCTTAGTACTAAGGAATCCTCTTGCTACAGCTGCAGGGAAGTAA
Protein
MTLEELSKIEEEEFSTGPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVKEMWTEVPRTGKGKKGKAVNKDKFISKMFLRGDSVILVLRNPLATAAGK
Summary
Description
Required for pre-mRNA splicing. Required for snRNP biogenesis (By similarity).
Subunit
Interacts with the SMN complex.
Similarity
Belongs to the snRNP core protein family.
Keywords
Complete proteome
mRNA processing
mRNA splicing
Nucleus
Reference proteome
Ribonucleoprotein
Feature
chain Small nuclear ribonucleoprotein Sm D2
Uniprot
S4PGW4
A0A212FIE6
A3QVV0
A0A194RU04
H9JMH0
A0A1E1WT76
+ More
A0A0N0P9V7 A0A2H1VJ16 A0A2A4IVA8 A0A0L7LPT5 A0A336MRD0 A0A336MLW6 A0A0A9Y193 A0A2P8YZ63 A0A224XPH3 A0A0P4VLF7 R4G8B8 T1HSQ6 A0A0C9RCH5 A0A2M3ZC74 A0A2M4AVD5 A0A2M4C3X1 A0A0L7RCB8 A0A154PHC9 A0A310SHI9 A0A2A3E611 A0A182FNY2 A0A1B6E1K1 E2AJ66 A0A087ZMS5 A0A026WWE6 A0A151JN53 E9J1R9 A0A151JVM5 E2C5K8 A0A158NJ78 K7ISL3 A0A067QWE7 A0A2J7Q7P2 A0A151I7C2 A0A232F041 A0A3L8D8T0 A0A2J7QXR4 E0W320 A0A2P8YEE6 E9HAP6 A0A023FF29 A0A023FT80 G3MLC9 A0A2R5LCJ9 A0A0L0BN20 B4JYD6 B7Q5F5 A0A0K8RBP5 A0A1B0BBX4 A0A3B0JDL6 A0A1A9UTL3 B4G3C0 A0A023FDV7 A0A1W4V437 B4MBN3 B4QYA4 B4KBP1 A0A3B0JCS8 Q298A9 A0A1B2AJC2 B4PVQ1 A0A0P9A149 B4I4E8 Q9VI10 A0A1Z5KX78 A0A224Z6N8 A0A131YEB2 L7M3G9 A0A131XDF6 A0A2P8YRW1 B3P2K1 A0A0M5J1Z8 A0A0P4ZYH9 A0A023EDG2 Q16UH6 A0A182T3Z9 A0A1B0CBQ6 A0A1I8P1B2 A0A1L8EGY0 T1P8H2 A0A0P5U499 A0A1A9W5Q0 A0A1B0DN66 A0A1B6GQP8 A0A182XH15 B4NHB4 A0A182NWL5 Q7Q2A9 A0A182IHA5 W8BV16 A0A034WSC1 A0A0C9QLU9 A0A0K8W6V6 A0A023ED47
A0A0N0P9V7 A0A2H1VJ16 A0A2A4IVA8 A0A0L7LPT5 A0A336MRD0 A0A336MLW6 A0A0A9Y193 A0A2P8YZ63 A0A224XPH3 A0A0P4VLF7 R4G8B8 T1HSQ6 A0A0C9RCH5 A0A2M3ZC74 A0A2M4AVD5 A0A2M4C3X1 A0A0L7RCB8 A0A154PHC9 A0A310SHI9 A0A2A3E611 A0A182FNY2 A0A1B6E1K1 E2AJ66 A0A087ZMS5 A0A026WWE6 A0A151JN53 E9J1R9 A0A151JVM5 E2C5K8 A0A158NJ78 K7ISL3 A0A067QWE7 A0A2J7Q7P2 A0A151I7C2 A0A232F041 A0A3L8D8T0 A0A2J7QXR4 E0W320 A0A2P8YEE6 E9HAP6 A0A023FF29 A0A023FT80 G3MLC9 A0A2R5LCJ9 A0A0L0BN20 B4JYD6 B7Q5F5 A0A0K8RBP5 A0A1B0BBX4 A0A3B0JDL6 A0A1A9UTL3 B4G3C0 A0A023FDV7 A0A1W4V437 B4MBN3 B4QYA4 B4KBP1 A0A3B0JCS8 Q298A9 A0A1B2AJC2 B4PVQ1 A0A0P9A149 B4I4E8 Q9VI10 A0A1Z5KX78 A0A224Z6N8 A0A131YEB2 L7M3G9 A0A131XDF6 A0A2P8YRW1 B3P2K1 A0A0M5J1Z8 A0A0P4ZYH9 A0A023EDG2 Q16UH6 A0A182T3Z9 A0A1B0CBQ6 A0A1I8P1B2 A0A1L8EGY0 T1P8H2 A0A0P5U499 A0A1A9W5Q0 A0A1B0DN66 A0A1B6GQP8 A0A182XH15 B4NHB4 A0A182NWL5 Q7Q2A9 A0A182IHA5 W8BV16 A0A034WSC1 A0A0C9QLU9 A0A0K8W6V6 A0A023ED47
Pubmed
23622113
22118469
26354079
19121390
26227816
25401762
+ More
29403074 27129103 20798317 24508170 21282665 21347285 20075255 24845553 28648823 30249741 20566863 21292972 22216098 26108605 17994087 15632085 17550304 10731132 12537572 12537569 10908584 18621711 17412918 28528879 28797301 26830274 25576852 28049606 24945155 17510324 25315136 12364791 14747013 17210077 24495485 25348373
29403074 27129103 20798317 24508170 21282665 21347285 20075255 24845553 28648823 30249741 20566863 21292972 22216098 26108605 17994087 15632085 17550304 10731132 12537572 12537569 10908584 18621711 17412918 28528879 28797301 26830274 25576852 28049606 24945155 17510324 25315136 12364791 14747013 17210077 24495485 25348373
EMBL
GAIX01006035
JAA86525.1
AGBW02008394
OWR53513.1
DQ868529
ABK42005.1
+ More
KQ459875 KPJ19601.1 BABH01016810 GDQN01000917 JAT90137.1 KQ459078 KPJ03815.1 ODYU01002840 SOQ40818.1 NWSH01007116 NWSH01003629 PCG63080.1 PCG66029.1 JTDY01000383 KOB77447.1 UFQT01002283 SSX33094.1 UFQT01001324 SSX30039.1 GBHO01020329 JAG23275.1 PYGN01000274 PSN49535.1 GFTR01002039 JAW14387.1 GDKW01001356 JAI55239.1 GAHY01001247 JAA76263.1 ACPB03006555 GBYB01004626 JAG74393.1 GGFM01005358 MBW26109.1 GGFK01011435 MBW44756.1 GGFJ01010808 MBW59949.1 KQ414617 KOC68381.1 KQ434899 KZC10884.1 KQ764145 OAD54593.1 KZ288363 PBC26924.1 GEDC01005517 JAS31781.1 GL439967 EFN66506.1 KK107078 EZA60400.1 KQ978858 KYN27926.1 GL767674 EFZ13177.1 KQ981703 KYN37440.1 GL452770 EFN76821.1 ADTU01017554 KK852869 KDR14658.1 NEVH01017441 PNF24590.1 KQ978411 KYM94099.1 NNAY01001464 OXU23887.1 QOIP01000011 RLU16701.1 NEVH01009372 PNF33377.1 DS235881 EEB20026.1 PYGN01000667 PSN42544.1 GL732612 EFX71229.1 GBBK01004330 JAC20152.1 GBBL01002334 JAC24986.1 JO842678 JO842679 JO842680 AEO34297.1 GGLE01003096 MBY07222.1 JRES01001623 KNC21387.1 CH916377 EDV90698.1 ABJB010745576 DS861322 EEC14077.1 GADI01005253 GEFM01003814 JAA68555.1 JAP71982.1 JXJN01011726 OUUW01000005 SPP80185.1 CH479179 EDW24302.1 GBBK01004331 JAC20151.1 CH940656 EDW58504.2 CM000364 EDX11861.1 CH933806 EDW14718.1 SPP80184.1 CM000070 EAL28046.3 KX531442 ANY27252.1 CM000160 EDW97860.2 CH902617 KPU80429.1 CH480821 EDW55091.1 AE014297 AY069315 AAL39460.1 GFJQ02007271 JAV99698.1 GFPF01011795 MAA22941.1 GEDV01011310 JAP77247.1 GACK01006469 JAA58565.1 GEFH01004960 JAP63621.1 PYGN01000402 PSN46977.1 CH954181 EDV48023.1 CP012526 ALC46811.1 GDIP01209555 JAJ13847.1 GAPW01006135 JAC07463.1 CH477622 EAT38161.1 AJWK01005764 GFDG01000809 JAV17990.1 KA644942 AFP59571.1 GDIP01118313 JAL85401.1 AJVK01017334 GECZ01005040 JAS64729.1 CH964272 EDW84590.2 AAAB01008978 EAA13684.2 APCN01002733 GAMC01003538 JAC03018.1 GAKP01002294 JAC56658.1 GBYB01004529 JAG74296.1 GDHF01014153 GDHF01005458 JAI38161.1 JAI46856.1 GAPW01006281 JAC07317.1
KQ459875 KPJ19601.1 BABH01016810 GDQN01000917 JAT90137.1 KQ459078 KPJ03815.1 ODYU01002840 SOQ40818.1 NWSH01007116 NWSH01003629 PCG63080.1 PCG66029.1 JTDY01000383 KOB77447.1 UFQT01002283 SSX33094.1 UFQT01001324 SSX30039.1 GBHO01020329 JAG23275.1 PYGN01000274 PSN49535.1 GFTR01002039 JAW14387.1 GDKW01001356 JAI55239.1 GAHY01001247 JAA76263.1 ACPB03006555 GBYB01004626 JAG74393.1 GGFM01005358 MBW26109.1 GGFK01011435 MBW44756.1 GGFJ01010808 MBW59949.1 KQ414617 KOC68381.1 KQ434899 KZC10884.1 KQ764145 OAD54593.1 KZ288363 PBC26924.1 GEDC01005517 JAS31781.1 GL439967 EFN66506.1 KK107078 EZA60400.1 KQ978858 KYN27926.1 GL767674 EFZ13177.1 KQ981703 KYN37440.1 GL452770 EFN76821.1 ADTU01017554 KK852869 KDR14658.1 NEVH01017441 PNF24590.1 KQ978411 KYM94099.1 NNAY01001464 OXU23887.1 QOIP01000011 RLU16701.1 NEVH01009372 PNF33377.1 DS235881 EEB20026.1 PYGN01000667 PSN42544.1 GL732612 EFX71229.1 GBBK01004330 JAC20152.1 GBBL01002334 JAC24986.1 JO842678 JO842679 JO842680 AEO34297.1 GGLE01003096 MBY07222.1 JRES01001623 KNC21387.1 CH916377 EDV90698.1 ABJB010745576 DS861322 EEC14077.1 GADI01005253 GEFM01003814 JAA68555.1 JAP71982.1 JXJN01011726 OUUW01000005 SPP80185.1 CH479179 EDW24302.1 GBBK01004331 JAC20151.1 CH940656 EDW58504.2 CM000364 EDX11861.1 CH933806 EDW14718.1 SPP80184.1 CM000070 EAL28046.3 KX531442 ANY27252.1 CM000160 EDW97860.2 CH902617 KPU80429.1 CH480821 EDW55091.1 AE014297 AY069315 AAL39460.1 GFJQ02007271 JAV99698.1 GFPF01011795 MAA22941.1 GEDV01011310 JAP77247.1 GACK01006469 JAA58565.1 GEFH01004960 JAP63621.1 PYGN01000402 PSN46977.1 CH954181 EDV48023.1 CP012526 ALC46811.1 GDIP01209555 JAJ13847.1 GAPW01006135 JAC07463.1 CH477622 EAT38161.1 AJWK01005764 GFDG01000809 JAV17990.1 KA644942 AFP59571.1 GDIP01118313 JAL85401.1 AJVK01017334 GECZ01005040 JAS64729.1 CH964272 EDW84590.2 AAAB01008978 EAA13684.2 APCN01002733 GAMC01003538 JAC03018.1 GAKP01002294 JAC56658.1 GBYB01004529 JAG74296.1 GDHF01014153 GDHF01005458 JAI38161.1 JAI46856.1 GAPW01006281 JAC07317.1
Proteomes
UP000007151
UP000053240
UP000005204
UP000053268
UP000218220
UP000037510
+ More
UP000245037 UP000015103 UP000053825 UP000076502 UP000242457 UP000069272 UP000000311 UP000005203 UP000053097 UP000078492 UP000078541 UP000008237 UP000005205 UP000002358 UP000027135 UP000235965 UP000078542 UP000215335 UP000279307 UP000009046 UP000000305 UP000037069 UP000001070 UP000001555 UP000092460 UP000268350 UP000078200 UP000008744 UP000192221 UP000008792 UP000000304 UP000009192 UP000001819 UP000002282 UP000007801 UP000001292 UP000000803 UP000008711 UP000092553 UP000008820 UP000075901 UP000092461 UP000095300 UP000095301 UP000091820 UP000092462 UP000076407 UP000007798 UP000075884 UP000007062 UP000075840
UP000245037 UP000015103 UP000053825 UP000076502 UP000242457 UP000069272 UP000000311 UP000005203 UP000053097 UP000078492 UP000078541 UP000008237 UP000005205 UP000002358 UP000027135 UP000235965 UP000078542 UP000215335 UP000279307 UP000009046 UP000000305 UP000037069 UP000001070 UP000001555 UP000092460 UP000268350 UP000078200 UP000008744 UP000192221 UP000008792 UP000000304 UP000009192 UP000001819 UP000002282 UP000007801 UP000001292 UP000000803 UP000008711 UP000092553 UP000008820 UP000075901 UP000092461 UP000095300 UP000095301 UP000091820 UP000092462 UP000076407 UP000007798 UP000075884 UP000007062 UP000075840
Interpro
Gene 3D
CDD
ProteinModelPortal
S4PGW4
A0A212FIE6
A3QVV0
A0A194RU04
H9JMH0
A0A1E1WT76
+ More
A0A0N0P9V7 A0A2H1VJ16 A0A2A4IVA8 A0A0L7LPT5 A0A336MRD0 A0A336MLW6 A0A0A9Y193 A0A2P8YZ63 A0A224XPH3 A0A0P4VLF7 R4G8B8 T1HSQ6 A0A0C9RCH5 A0A2M3ZC74 A0A2M4AVD5 A0A2M4C3X1 A0A0L7RCB8 A0A154PHC9 A0A310SHI9 A0A2A3E611 A0A182FNY2 A0A1B6E1K1 E2AJ66 A0A087ZMS5 A0A026WWE6 A0A151JN53 E9J1R9 A0A151JVM5 E2C5K8 A0A158NJ78 K7ISL3 A0A067QWE7 A0A2J7Q7P2 A0A151I7C2 A0A232F041 A0A3L8D8T0 A0A2J7QXR4 E0W320 A0A2P8YEE6 E9HAP6 A0A023FF29 A0A023FT80 G3MLC9 A0A2R5LCJ9 A0A0L0BN20 B4JYD6 B7Q5F5 A0A0K8RBP5 A0A1B0BBX4 A0A3B0JDL6 A0A1A9UTL3 B4G3C0 A0A023FDV7 A0A1W4V437 B4MBN3 B4QYA4 B4KBP1 A0A3B0JCS8 Q298A9 A0A1B2AJC2 B4PVQ1 A0A0P9A149 B4I4E8 Q9VI10 A0A1Z5KX78 A0A224Z6N8 A0A131YEB2 L7M3G9 A0A131XDF6 A0A2P8YRW1 B3P2K1 A0A0M5J1Z8 A0A0P4ZYH9 A0A023EDG2 Q16UH6 A0A182T3Z9 A0A1B0CBQ6 A0A1I8P1B2 A0A1L8EGY0 T1P8H2 A0A0P5U499 A0A1A9W5Q0 A0A1B0DN66 A0A1B6GQP8 A0A182XH15 B4NHB4 A0A182NWL5 Q7Q2A9 A0A182IHA5 W8BV16 A0A034WSC1 A0A0C9QLU9 A0A0K8W6V6 A0A023ED47
A0A0N0P9V7 A0A2H1VJ16 A0A2A4IVA8 A0A0L7LPT5 A0A336MRD0 A0A336MLW6 A0A0A9Y193 A0A2P8YZ63 A0A224XPH3 A0A0P4VLF7 R4G8B8 T1HSQ6 A0A0C9RCH5 A0A2M3ZC74 A0A2M4AVD5 A0A2M4C3X1 A0A0L7RCB8 A0A154PHC9 A0A310SHI9 A0A2A3E611 A0A182FNY2 A0A1B6E1K1 E2AJ66 A0A087ZMS5 A0A026WWE6 A0A151JN53 E9J1R9 A0A151JVM5 E2C5K8 A0A158NJ78 K7ISL3 A0A067QWE7 A0A2J7Q7P2 A0A151I7C2 A0A232F041 A0A3L8D8T0 A0A2J7QXR4 E0W320 A0A2P8YEE6 E9HAP6 A0A023FF29 A0A023FT80 G3MLC9 A0A2R5LCJ9 A0A0L0BN20 B4JYD6 B7Q5F5 A0A0K8RBP5 A0A1B0BBX4 A0A3B0JDL6 A0A1A9UTL3 B4G3C0 A0A023FDV7 A0A1W4V437 B4MBN3 B4QYA4 B4KBP1 A0A3B0JCS8 Q298A9 A0A1B2AJC2 B4PVQ1 A0A0P9A149 B4I4E8 Q9VI10 A0A1Z5KX78 A0A224Z6N8 A0A131YEB2 L7M3G9 A0A131XDF6 A0A2P8YRW1 B3P2K1 A0A0M5J1Z8 A0A0P4ZYH9 A0A023EDG2 Q16UH6 A0A182T3Z9 A0A1B0CBQ6 A0A1I8P1B2 A0A1L8EGY0 T1P8H2 A0A0P5U499 A0A1A9W5Q0 A0A1B0DN66 A0A1B6GQP8 A0A182XH15 B4NHB4 A0A182NWL5 Q7Q2A9 A0A182IHA5 W8BV16 A0A034WSC1 A0A0C9QLU9 A0A0K8W6V6 A0A023ED47
PDB
3JB9
E-value=1.01692e-31,
Score=334
Ontologies
KEGG
GO
PANTHER
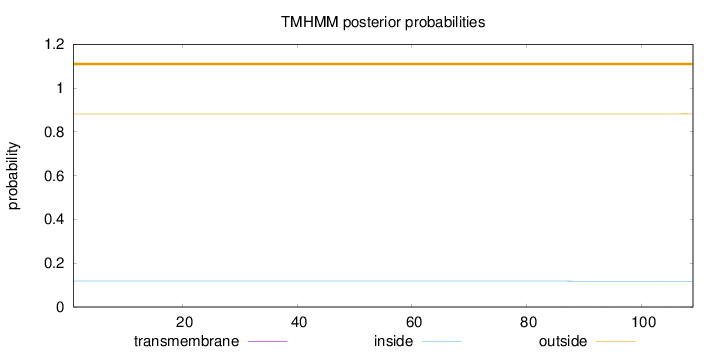
Topology
Subcellular location
Nucleus
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01042
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.11839
outside
1 - 109
Population Genetic Test Statistics
Pi
296.337896
Theta
202.199577
Tajima's D
1.557543
CLR
0
CSRT
0.799110044497775
Interpretation
Uncertain
