Pre Gene Modal
BGIBMGA010829
Annotation
PREDICTED:_glycerol-3-phosphate_acyltransferase_1?_mitochondrial_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.399
Sequence
CDS
ATGGATGCTTTGCCGCGCGTCCAACAACTTCGTGATAATTATGACTTGCAAAAACTAGTGATTAGAGAAATTCGTGAAAAAGTGTTAGTCAAGATTAGCCCTGGAAGCCTGTCGTGGAACTTGACCGGTGGTGATGTAAAACTGAAAGCTGACGTGATGGAAACGATGATCAGCGTGTGGACTGTGGTGGAGCATGTGAGGGTAGCCAGTGGTGCCGCTGAAGTGGCCATCTTCACTGCAATACTCTACTGGTTTTTCACTTCCAGCCGGGTAGCGGATATGCTAGAAGTGCTTGGAGAAAGGGTCGGGGGATGGTGCGGACGAGAGTCTGGCTCGCTGCGGCAGGCAGTGCGTGCCCGCAGGAGACAACAAAATGCCGACATGTATGCCAAGCTCGACGCTGACGTTGCTGTTCGTACGACGTCTCTGTATAGAGTGAAAGAGAGCCCCGTGGTTCCAGCGCCGCAACCGGAGACAAGGCCCCCAGCTGGACTGGCGTGCGGCCGGTGCGCTCCTGTTTCAAGGGATTCCTGGCACGACCCGAAGATCGAAGACTCCGGTACCGTGATCAACGTGTTAGATATAGGAAGACAGAACTTTGCGAATAGTGGCGTTATCTCAAGATATTTCTGTGATCTAGCGCAATGTTTCAATCTTGTCAAGTTCCAGTACAAGGATGTTATTCCAGATGTGCAAAAGGATCCAGCTTACATAAAAGCTATCGAAGACACAACCGAAGAAGAACTAAAAAAGACTGATTTGAACCTAGACGAAAAGGGAGATGGAGATAATCGAAACAGCGCCGGCTATCTGACCGTGAAGAAACGCGTCGAAGCCAGAGCGAAAAAAGTCTTGACCGATATTAGTTCGGCCATGTCTTACAATATACTAAGAGTGGTGGCGTGGCTGACGCACAAGGCGATCCGTCGCGTGGCGGGCGGCGGGTGCGGCACGCGCGCGGGCTCGGTGGCGCGGCTGCGGCGCGCGACGGCGGCGGGGCTGCCGCTCGTGTTCGTGCCGCTGCACCGCTCGCACTTCGACTACATCCTCGTCACCTTCGCGCTCTACCTCACCGGGCTGCGGCCGCCGCTCGTGGCCGCCGGGGACAACATGAGGATACCCTTCTTTGGGTGGATACTGCGCGGCTGCGGCGCTTTTTACATCAGACGAAGAGTTGACGGTTCCGAGCACAACGGCGACCCCGTCTATAAAGCAGCTTTAAGATCTTACATCATAAACAGTTTGGCGGCCAACAACAATCTGGAGTTCTTCATTGAAGGCGGCCGGACTAGGACCGGGAAGCCGCAGCCTCCCAAAGCTGGAATCCTGTCCGTGATAATGGATGCGTATTTGGATGGCACCATAGACGACGCCCTCCTAGTCCCGGTCACGTTGAACTACGACAAGTTGGTCGACGGTAACTTCGTGAGGGAACAACTCGGCATGCCCAAGCAGATGGAGACGTTCTGGTCGGCCCTCAGAGGGATCTGGAGGACAATGAACACTAATCACGGCTCTATAAGGGTCGATTTCAATCAACCCATCTCTTTGAAGGAATTGGTGCAATCTTTCAAAAAGTATAATCACTTCAAAGCTCCCATCGAAGCGCCTCTTGGTCCTGCCGATGACAAATTTTCTTCGGATCTGGACCGGAGACTGTTGTACAACCACAGCCACTCGAGCTTGTACGGCGACGACGTCAGCACTGACCACAAGATGATGGTCGAGGCTATTGGGAGACATATTGTTTATGAGGGTGCTCAGGCCACGGCGGTGATGTGCACGAACGTGGTGTCGTACGTGCTGCTGACGGAGGCGCGCGGCGGCGCGGGCGTGCGGCGCGCCGTGCGGGCTCGCGGCGCCGCGCTGCGGGCGGAGGGCCGGGACCTCGCCTACCGCGCCGACCCCGACCGCTGTCTGGAGCACGCGTTGGAAATGCTCGGGCCCCTTGTGCAGCGAGAGCGGAGAACCGAAACACTTCGTCCTACTAAATCGATAGCCGCCGAGCTGGAACTCTCGTATTACGCCAACACTCTGGTCGCGCACTACGCCGCGCCCGCTATCGTCGCCACGGCCCTCGAGAGTATAGTGTGTCGGCCGGATGCGGATGGGGACACGGTGCGGCATCAGGAGCTGCTGGGCTCGGCGCTGCAGCTGTACGAGGTGCTGAGCGAGGAGCTTATCCTGTGCGCGCCGTGCCACACCATCCACGAGACGCTGTTGCGGGCGCTCGACCAGCTCCTGGCCAGCGGGGTCCTTGTTAATGCAGAGCGCACTGACGGCTTAGAGGAAGAGAAGTGGTCTCGTCGATTTGCCAAGGCTATAGATGACGAGGAAGACGACGGCTACGAGCGACCCGACCCGACCTTGAGAATCAAGTACATCATTCAGAAAACTCCTGAAGCGATCGCTGAGAGACGTCGCCTGCTGCTGACCCTGCGTCCCCTCATCGAGTCGTACAGCGCGTCGTGCCGGCACCTCGCCCCCGGGACCGTCAAGTCGGTCGTCAAGGCGACCATCGACGGTCTCGTCCACGACTACACTACTAACAAGATGCCCTACGGCGCGGCCGTCTCTAGCGACGCGGTCAGGAACTGCCTGCGTCTCCTGAGGCAATGGGGCGTCATAGAAATGTACACGGACAACAAGGAACGGAAGATAAGGCTGAGGCCTCCCTACGACGACGCGCAGAAACTGAATGACATCTGCTCCCACATCTACAAGTTCAATGTCGACTCGCCTTTGCTCAGGGAGGAAAGCGCCACGTGA
Protein
MDALPRVQQLRDNYDLQKLVIREIREKVLVKISPGSLSWNLTGGDVKLKADVMETMISVWTVVEHVRVASGAAEVAIFTAILYWFFTSSRVADMLEVLGERVGGWCGRESGSLRQAVRARRRQQNADMYAKLDADVAVRTTSLYRVKESPVVPAPQPETRPPAGLACGRCAPVSRDSWHDPKIEDSGTVINVLDIGRQNFANSGVISRYFCDLAQCFNLVKFQYKDVIPDVQKDPAYIKAIEDTTEEELKKTDLNLDEKGDGDNRNSAGYLTVKKRVEARAKKVLTDISSAMSYNILRVVAWLTHKAIRRVAGGGCGTRAGSVARLRRATAAGLPLVFVPLHRSHFDYILVTFALYLTGLRPPLVAAGDNMRIPFFGWILRGCGAFYIRRRVDGSEHNGDPVYKAALRSYIINSLAANNNLEFFIEGGRTRTGKPQPPKAGILSVIMDAYLDGTIDDALLVPVTLNYDKLVDGNFVREQLGMPKQMETFWSALRGIWRTMNTNHGSIRVDFNQPISLKELVQSFKKYNHFKAPIEAPLGPADDKFSSDLDRRLLYNHSHSSLYGDDVSTDHKMMVEAIGRHIVYEGAQATAVMCTNVVSYVLLTEARGGAGVRRAVRARGAALRAEGRDLAYRADPDRCLEHALEMLGPLVQRERRTETLRPTKSIAAELELSYYANTLVAHYAAPAIVATALESIVCRPDADGDTVRHQELLGSALQLYEVLSEELILCAPCHTIHETLLRALDQLLASGVLVNAERTDGLEEEKWSRRFAKAIDDEEDDGYERPDPTLRIKYIIQKTPEAIAERRRLLLTLRPLIESYSASCRHLAPGTVKSVVKATIDGLVHDYTTNKMPYGAAVSSDAVRNCLRLLRQWGVIEMYTDNKERKIRLRPPYDDAQKLNDICSHIYKFNVDSPLLREESAT
Summary
Similarity
Belongs to the GPAT/DAPAT family.
Uniprot
H9JMS6
S4PSC7
A0A0N1IBW6
A0A3S2LD17
A0A212F0H7
D6X1Q9
+ More
A0A0T6B997 A0A2J7RR58 E2A4R3 A0A1Y1MQ89 A0A026W9I9 A0A2J7RR57 A0A1B6LWG5 N6SYL6 U4UFK2 A0A1Y1MRY0 A0A0C9RZT5 A0A0C9S261 A0A2J7RR53 A0A154P3H8 A0A2J7RR73 A0A232FGD6 A0A151IKB1 F4W9X6 K7IU60 A0A0L7QYS6 A0A088AF61 E9I9J5 A0A195EEU6 A0A1L8DYV3 A0A0A9YUL9 A0A0A9Z0H8 A0A158ND30 A0A2A3EPK4 A0A336MI00 A0A0A9YQJ3 A0A034VZ72 A0A034W0R9 A0A182RZD1 A0A182YG68 A0A195BQ95 A0A224XK13 A0A151WKP2 E0VA08 A0A0K8VKD6 A0A0K8W9X9 A0A182NCJ8 A0A1B0CQF3 A0A1I8MC63 T1PE91 A0A182LSP3 A0A2M4CZY5 Q173M5 A0A1I8MC68 A0A1S4FFL0 A0A1I8MC65 A0A0L0CH59 A0A182QUZ3 B3M018 A0A182JBI1 B4NJQ2 A0A182V8R5 A0A182WVG5 A0A182I944 A0A0P9CCA1 A0A182FWE1 A0A1B6DIG6 A0A1B6E8Z5 A0A195EZ77 A0A0V0G495 A0A0N8P1R9 A0A182PTS5 A0A0K8SK80 B4JEQ4 A0A336MYD6 A0A240SX65 A0A240SWK7 A0A0Q9WYI1 A0A1I8P7X6 A0A2M4BDN6 A0A182KXX2 Q7QAI3 B4K9R0 W5JHU5 A0A2M4BDK7 A0A240SX72 A0A240SWK6
A0A0T6B997 A0A2J7RR58 E2A4R3 A0A1Y1MQ89 A0A026W9I9 A0A2J7RR57 A0A1B6LWG5 N6SYL6 U4UFK2 A0A1Y1MRY0 A0A0C9RZT5 A0A0C9S261 A0A2J7RR53 A0A154P3H8 A0A2J7RR73 A0A232FGD6 A0A151IKB1 F4W9X6 K7IU60 A0A0L7QYS6 A0A088AF61 E9I9J5 A0A195EEU6 A0A1L8DYV3 A0A0A9YUL9 A0A0A9Z0H8 A0A158ND30 A0A2A3EPK4 A0A336MI00 A0A0A9YQJ3 A0A034VZ72 A0A034W0R9 A0A182RZD1 A0A182YG68 A0A195BQ95 A0A224XK13 A0A151WKP2 E0VA08 A0A0K8VKD6 A0A0K8W9X9 A0A182NCJ8 A0A1B0CQF3 A0A1I8MC63 T1PE91 A0A182LSP3 A0A2M4CZY5 Q173M5 A0A1I8MC68 A0A1S4FFL0 A0A1I8MC65 A0A0L0CH59 A0A182QUZ3 B3M018 A0A182JBI1 B4NJQ2 A0A182V8R5 A0A182WVG5 A0A182I944 A0A0P9CCA1 A0A182FWE1 A0A1B6DIG6 A0A1B6E8Z5 A0A195EZ77 A0A0V0G495 A0A0N8P1R9 A0A182PTS5 A0A0K8SK80 B4JEQ4 A0A336MYD6 A0A240SX65 A0A240SWK7 A0A0Q9WYI1 A0A1I8P7X6 A0A2M4BDN6 A0A182KXX2 Q7QAI3 B4K9R0 W5JHU5 A0A2M4BDK7 A0A240SX72 A0A240SWK6
Pubmed
EMBL
BABH01016802
BABH01016803
GAIX01013973
JAA78587.1
KQ459078
KPJ03819.1
+ More
RSAL01000221 RVE44058.1 AGBW02011075 OWR47249.1 KQ971371 EFA09975.1 LJIG01009262 KRT83431.1 NEVH01000611 PNF43319.1 GL436716 EFN71597.1 GEZM01026093 JAV87248.1 KK107321 QOIP01000002 EZA52710.1 RLU25366.1 PNF43320.1 GEBQ01011958 JAT28019.1 APGK01058794 KB741292 ENN70308.1 KB632334 ERL92764.1 GEZM01026095 JAV87245.1 GBYB01014990 JAG84757.1 GBYB01014991 JAG84758.1 PNF43318.1 KQ434803 KZC05904.1 PNF43317.1 NNAY01000282 OXU29518.1 KQ977234 KYN04710.1 GL888033 EGI69112.1 AAZX01002636 KQ414688 KOC63702.1 GL761812 EFZ22772.1 KQ979039 KYN23327.1 GFDF01002629 JAV11455.1 GBHO01008288 GBRD01005011 JAG35316.1 JAG60810.1 GBHO01008289 JAG35315.1 ADTU01000889 ADTU01000890 ADTU01000891 ADTU01000892 KZ288199 PBC33630.1 UFQS01001326 UFQT01001326 SSX10364.1 SSX30052.1 GBHO01008287 GBRD01005014 GBRD01005012 JAG35317.1 JAG60807.1 GAKP01011267 GAKP01011263 GAKP01011261 JAC47689.1 GAKP01011265 GAKP01011259 GAKP01011257 GAKP01011255 JAC47695.1 KQ976423 KYM88746.1 GFTR01008055 JAW08371.1 KQ983012 KYQ48377.1 DS235004 EEB10214.1 GDHF01013284 JAI39030.1 GDHF01004427 JAI47887.1 AJWK01023485 AJWK01023486 AJWK01023487 AJWK01023488 AJWK01023489 KA647097 AFP61726.1 AXCM01000095 GGFL01006661 MBW70839.1 CH477419 EAT41268.1 JRES01000409 KNC31537.1 AXCN02001185 CH902617 EDV44208.1 CH964272 EDW85014.1 APCN01003173 KPU80693.1 GEDC01011805 JAS25493.1 GEDC01002898 JAS34400.1 KQ981909 KYN33212.1 GECL01003263 JAP02861.1 KPU80692.1 GBRD01012141 JAG53683.1 CH916369 EDV93185.1 UFQS01003970 UFQT01003970 SSX16069.1 SSX35402.1 CH933806 KRG01055.1 GGFJ01002014 MBW51155.1 AAAB01008888 EAA08751.4 EDW14535.1 ADMH02001205 ETN63691.1 GGFJ01001984 MBW51125.1
RSAL01000221 RVE44058.1 AGBW02011075 OWR47249.1 KQ971371 EFA09975.1 LJIG01009262 KRT83431.1 NEVH01000611 PNF43319.1 GL436716 EFN71597.1 GEZM01026093 JAV87248.1 KK107321 QOIP01000002 EZA52710.1 RLU25366.1 PNF43320.1 GEBQ01011958 JAT28019.1 APGK01058794 KB741292 ENN70308.1 KB632334 ERL92764.1 GEZM01026095 JAV87245.1 GBYB01014990 JAG84757.1 GBYB01014991 JAG84758.1 PNF43318.1 KQ434803 KZC05904.1 PNF43317.1 NNAY01000282 OXU29518.1 KQ977234 KYN04710.1 GL888033 EGI69112.1 AAZX01002636 KQ414688 KOC63702.1 GL761812 EFZ22772.1 KQ979039 KYN23327.1 GFDF01002629 JAV11455.1 GBHO01008288 GBRD01005011 JAG35316.1 JAG60810.1 GBHO01008289 JAG35315.1 ADTU01000889 ADTU01000890 ADTU01000891 ADTU01000892 KZ288199 PBC33630.1 UFQS01001326 UFQT01001326 SSX10364.1 SSX30052.1 GBHO01008287 GBRD01005014 GBRD01005012 JAG35317.1 JAG60807.1 GAKP01011267 GAKP01011263 GAKP01011261 JAC47689.1 GAKP01011265 GAKP01011259 GAKP01011257 GAKP01011255 JAC47695.1 KQ976423 KYM88746.1 GFTR01008055 JAW08371.1 KQ983012 KYQ48377.1 DS235004 EEB10214.1 GDHF01013284 JAI39030.1 GDHF01004427 JAI47887.1 AJWK01023485 AJWK01023486 AJWK01023487 AJWK01023488 AJWK01023489 KA647097 AFP61726.1 AXCM01000095 GGFL01006661 MBW70839.1 CH477419 EAT41268.1 JRES01000409 KNC31537.1 AXCN02001185 CH902617 EDV44208.1 CH964272 EDW85014.1 APCN01003173 KPU80693.1 GEDC01011805 JAS25493.1 GEDC01002898 JAS34400.1 KQ981909 KYN33212.1 GECL01003263 JAP02861.1 KPU80692.1 GBRD01012141 JAG53683.1 CH916369 EDV93185.1 UFQS01003970 UFQT01003970 SSX16069.1 SSX35402.1 CH933806 KRG01055.1 GGFJ01002014 MBW51155.1 AAAB01008888 EAA08751.4 EDW14535.1 ADMH02001205 ETN63691.1 GGFJ01001984 MBW51125.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000007266
UP000235965
+ More
UP000000311 UP000053097 UP000279307 UP000019118 UP000030742 UP000076502 UP000215335 UP000078542 UP000007755 UP000002358 UP000053825 UP000005203 UP000078492 UP000005205 UP000242457 UP000075900 UP000076408 UP000078540 UP000075809 UP000009046 UP000075884 UP000092461 UP000095301 UP000075883 UP000008820 UP000037069 UP000075886 UP000007801 UP000075880 UP000007798 UP000075903 UP000076407 UP000075840 UP000069272 UP000078541 UP000075885 UP000001070 UP000092445 UP000092443 UP000009192 UP000095300 UP000075882 UP000007062 UP000000673
UP000000311 UP000053097 UP000279307 UP000019118 UP000030742 UP000076502 UP000215335 UP000078542 UP000007755 UP000002358 UP000053825 UP000005203 UP000078492 UP000005205 UP000242457 UP000075900 UP000076408 UP000078540 UP000075809 UP000009046 UP000075884 UP000092461 UP000095301 UP000075883 UP000008820 UP000037069 UP000075886 UP000007801 UP000075880 UP000007798 UP000075903 UP000076407 UP000075840 UP000069272 UP000078541 UP000075885 UP000001070 UP000092445 UP000092443 UP000009192 UP000095300 UP000075882 UP000007062 UP000000673
PRIDE
Interpro
ProteinModelPortal
H9JMS6
S4PSC7
A0A0N1IBW6
A0A3S2LD17
A0A212F0H7
D6X1Q9
+ More
A0A0T6B997 A0A2J7RR58 E2A4R3 A0A1Y1MQ89 A0A026W9I9 A0A2J7RR57 A0A1B6LWG5 N6SYL6 U4UFK2 A0A1Y1MRY0 A0A0C9RZT5 A0A0C9S261 A0A2J7RR53 A0A154P3H8 A0A2J7RR73 A0A232FGD6 A0A151IKB1 F4W9X6 K7IU60 A0A0L7QYS6 A0A088AF61 E9I9J5 A0A195EEU6 A0A1L8DYV3 A0A0A9YUL9 A0A0A9Z0H8 A0A158ND30 A0A2A3EPK4 A0A336MI00 A0A0A9YQJ3 A0A034VZ72 A0A034W0R9 A0A182RZD1 A0A182YG68 A0A195BQ95 A0A224XK13 A0A151WKP2 E0VA08 A0A0K8VKD6 A0A0K8W9X9 A0A182NCJ8 A0A1B0CQF3 A0A1I8MC63 T1PE91 A0A182LSP3 A0A2M4CZY5 Q173M5 A0A1I8MC68 A0A1S4FFL0 A0A1I8MC65 A0A0L0CH59 A0A182QUZ3 B3M018 A0A182JBI1 B4NJQ2 A0A182V8R5 A0A182WVG5 A0A182I944 A0A0P9CCA1 A0A182FWE1 A0A1B6DIG6 A0A1B6E8Z5 A0A195EZ77 A0A0V0G495 A0A0N8P1R9 A0A182PTS5 A0A0K8SK80 B4JEQ4 A0A336MYD6 A0A240SX65 A0A240SWK7 A0A0Q9WYI1 A0A1I8P7X6 A0A2M4BDN6 A0A182KXX2 Q7QAI3 B4K9R0 W5JHU5 A0A2M4BDK7 A0A240SX72 A0A240SWK6
A0A0T6B997 A0A2J7RR58 E2A4R3 A0A1Y1MQ89 A0A026W9I9 A0A2J7RR57 A0A1B6LWG5 N6SYL6 U4UFK2 A0A1Y1MRY0 A0A0C9RZT5 A0A0C9S261 A0A2J7RR53 A0A154P3H8 A0A2J7RR73 A0A232FGD6 A0A151IKB1 F4W9X6 K7IU60 A0A0L7QYS6 A0A088AF61 E9I9J5 A0A195EEU6 A0A1L8DYV3 A0A0A9YUL9 A0A0A9Z0H8 A0A158ND30 A0A2A3EPK4 A0A336MI00 A0A0A9YQJ3 A0A034VZ72 A0A034W0R9 A0A182RZD1 A0A182YG68 A0A195BQ95 A0A224XK13 A0A151WKP2 E0VA08 A0A0K8VKD6 A0A0K8W9X9 A0A182NCJ8 A0A1B0CQF3 A0A1I8MC63 T1PE91 A0A182LSP3 A0A2M4CZY5 Q173M5 A0A1I8MC68 A0A1S4FFL0 A0A1I8MC65 A0A0L0CH59 A0A182QUZ3 B3M018 A0A182JBI1 B4NJQ2 A0A182V8R5 A0A182WVG5 A0A182I944 A0A0P9CCA1 A0A182FWE1 A0A1B6DIG6 A0A1B6E8Z5 A0A195EZ77 A0A0V0G495 A0A0N8P1R9 A0A182PTS5 A0A0K8SK80 B4JEQ4 A0A336MYD6 A0A240SX65 A0A240SWK7 A0A0Q9WYI1 A0A1I8P7X6 A0A2M4BDN6 A0A182KXX2 Q7QAI3 B4K9R0 W5JHU5 A0A2M4BDK7 A0A240SX72 A0A240SWK6
Ontologies
PATHWAY
GO
GO:0044255
GO:0008374
GO:0016021
GO:0019432
GO:0004366
GO:0008654
GO:0031966
GO:0006631
GO:0006072
GO:0015991
GO:0046961
GO:0033180
GO:0046933
GO:0102420
GO:0005886
GO:0005783
GO:0005739
GO:0035324
GO:0034587
GO:0045169
GO:0008152
GO:0016746
GO:0006886
GO:0016192
GO:0030126
GO:0030117
GO:0006418
GO:0003707
GO:0006281
GO:0006259
PANTHER
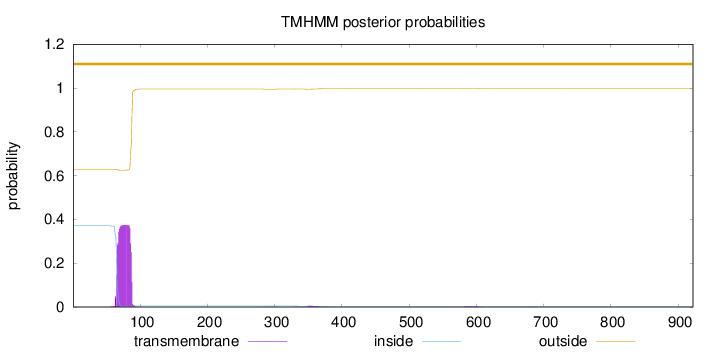
Topology
Length:
922
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.37041999999997
Exp number, first 60 AAs:
0.00763
Total prob of N-in:
0.37100
outside
1 - 922
Population Genetic Test Statistics
Pi
242.073342
Theta
166.664101
Tajima's D
1.281567
CLR
0.278432
CSRT
0.730513474326284
Interpretation
Uncertain
