Pre Gene Modal
BGIBMGA010724
Annotation
PREDICTED:_charged_multivesicular_body_protein_3_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.75 Nuclear Reliability : 2.365
Sequence
CDS
ATGGGCCTTTTCGGTAAATCCCCAGAGCGCAACCCTAAAGAAATGGTAAATGAATGGTCGCATAAACTTCGCAAAGAAGGATACAATTTGGACAGACAAGTTAGAGCTATCCAAAGAGAAGAAGAAAAAATTAAACGATCACTGAAAGAAGCAGCTACTAAAAACGATAAACAAGTATGCACTATACTAGCAAAAGAAATCATCAGATCCAGGAAAGCAATCAATAAAATATACACAAGTAAAGCACATCTCAACTCAGTACAATTACAAATGAAGAATCAATTGGCTACCTTAAGAGTTGCTGGATCGTTGCAGAAGTCAACAGAGGTGATGCAAGCAATGCAAGCTCTGTTACGTCTCCCAGAAGTAGCGGCTACGATGCAGGAGCTGAGCAAGGAAATGATGAAAGCTGGTATCATAGAGGAGATGCTGGACGAAACCATGTCCGGGATGGAAGATGAAGAGGAAATGGAAGAAGCAGCGCAAAGTGAAGTTGATAAGGTTCTATGGGAATTAACTCAAGGAAAATTAGGAGAAGCGCCGGCACCGCCCACATCTGTCGGAGCACCGTCCACCAGCCAACAAGAAGAGGAACCAGTCCCGGAGCCTGACGAAAGCGAACTGGACGAAATGCAGTCTAGATTAGAATCATTGCGGTCTTAG
Protein
MGLFGKSPERNPKEMVNEWSHKLRKEGYNLDRQVRAIQREEEKIKRSLKEAATKNDKQVCTILAKEIIRSRKAINKIYTSKAHLNSVQLQMKNQLATLRVAGSLQKSTEVMQAMQALLRLPEVAATMQELSKEMMKAGIIEEMLDETMSGMEDEEEMEEAAQSEVDKVLWELTQGKLGEAPAPPTSVGAPSTSQQEEEPVPEPDESELDEMQSRLESLRS
Summary
Uniprot
A0A1E1W5M1
A0A194RQL6
A0A1V0JHV4
A0A0N1PF08
I4DKD7
A0A2A4JUK8
+ More
A0A212EIR8 A0A2M4BZ49 A0A2M4AKR4 A0A2M3Z7N0 A0A1Y1KT70 A0A067RG49 A0A1W4WMF6 W5J6Q6 A0A182FL05 A0A182KFS0 A0A182NSJ0 A0A182VS48 A0A2J7PYU5 T1DG90 A0A182R892 A0A182QQS7 D6X057 N6TGA5 A0A182YLL9 Q06DJ7 A0A182TWV2 A0A182WW00 Q7QH42 A0A182P088 A0A182UQ78 A0A084VDU2 A0A0N1ITF9 A0A2M4BZQ8 A0A1L8DVS5 U5EX10 A0A182MPT5 A0A088AF24 A0A195FKQ0 E2AML3 A0A195BAQ3 A0A158NU97 F4WR85 Q171V6 A0A151JCE6 A0A023EK15 A0A069DQ93 A0A0J7KPB8 B4PTQ5 A0A151IQK9 B4K985 A0A0M4EP29 A0A336M1F5 Q295E5 A0A232FN59 A0A0P4VL65 T1IEU2 B4GM56 A0A182INF8 A0A0A1WRL9 A0A0C9QDW5 W8CBW2 A0A026WRE1 A0A0K8WHU3 A0A034W6U4 K7INY3 A0A1L8DVM1 B4LYX0 E2B9G8 Q9VN02 A0A3B0KSV6 A0A1W4VL71 B4QUZ9 B4I3L8 B3P1Y7 A0A0A9WH18 T1GBQ4 B0WAJ4 B4JV71 B3LXZ9 A0A2M4AMZ7 A0A023F592 A0A0V0G921 A0A0K8UAL6 A0A1Q3FF52 B4N924 A0A1B6JEX8 A0A1Q3FBK3 A0A1I8NV40 A0A1B6F0T6 A0A154PBN0 E0VVA7 A0A1I8MIU2 A0A1J1J485 A0A0L0CJM6 A0A162PB75 A0A146LBE1 A0A1B0FFU9 A0A1A9YQ12 A0A293MW15
A0A212EIR8 A0A2M4BZ49 A0A2M4AKR4 A0A2M3Z7N0 A0A1Y1KT70 A0A067RG49 A0A1W4WMF6 W5J6Q6 A0A182FL05 A0A182KFS0 A0A182NSJ0 A0A182VS48 A0A2J7PYU5 T1DG90 A0A182R892 A0A182QQS7 D6X057 N6TGA5 A0A182YLL9 Q06DJ7 A0A182TWV2 A0A182WW00 Q7QH42 A0A182P088 A0A182UQ78 A0A084VDU2 A0A0N1ITF9 A0A2M4BZQ8 A0A1L8DVS5 U5EX10 A0A182MPT5 A0A088AF24 A0A195FKQ0 E2AML3 A0A195BAQ3 A0A158NU97 F4WR85 Q171V6 A0A151JCE6 A0A023EK15 A0A069DQ93 A0A0J7KPB8 B4PTQ5 A0A151IQK9 B4K985 A0A0M4EP29 A0A336M1F5 Q295E5 A0A232FN59 A0A0P4VL65 T1IEU2 B4GM56 A0A182INF8 A0A0A1WRL9 A0A0C9QDW5 W8CBW2 A0A026WRE1 A0A0K8WHU3 A0A034W6U4 K7INY3 A0A1L8DVM1 B4LYX0 E2B9G8 Q9VN02 A0A3B0KSV6 A0A1W4VL71 B4QUZ9 B4I3L8 B3P1Y7 A0A0A9WH18 T1GBQ4 B0WAJ4 B4JV71 B3LXZ9 A0A2M4AMZ7 A0A023F592 A0A0V0G921 A0A0K8UAL6 A0A1Q3FF52 B4N924 A0A1B6JEX8 A0A1Q3FBK3 A0A1I8NV40 A0A1B6F0T6 A0A154PBN0 E0VVA7 A0A1I8MIU2 A0A1J1J485 A0A0L0CJM6 A0A162PB75 A0A146LBE1 A0A1B0FFU9 A0A1A9YQ12 A0A293MW15
Pubmed
26354079
22651552
22118469
28004739
24845553
20920257
+ More
23761445 18362917 19820115 23537049 25244985 17244545 12364791 14747013 17210077 24438588 20798317 21347285 21719571 17510324 24945155 26483478 26334808 17994087 17550304 15632085 28648823 27129103 25830018 24495485 24508170 30249741 25348373 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 25474469 20566863 25315136 26108605 26823975
23761445 18362917 19820115 23537049 25244985 17244545 12364791 14747013 17210077 24438588 20798317 21347285 21719571 17510324 24945155 26483478 26334808 17994087 17550304 15632085 28648823 27129103 25830018 24495485 24508170 30249741 25348373 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 25474469 20566863 25315136 26108605 26823975
EMBL
GDQN01008800
JAT82254.1
KQ459875
KPJ19605.1
KY694524
ODYU01000194
+ More
ARD08865.1 SOQ34575.1 KQ459078 KPJ03821.1 AK401755 BAM18377.1 NWSH01000595 PCG75456.1 AGBW02014588 OWR41375.1 GGFJ01009199 MBW58340.1 GGFK01008052 MBW41373.1 GGFM01003760 MBW24511.1 GEZM01074367 JAV64583.1 KK852496 KDR22737.1 ADMH02002130 ETN58540.1 NEVH01020342 PNF21511.1 GAMD01002851 JAA98739.1 AXCN02000234 KQ971371 EFA10075.1 APGK01028591 KB740694 KI208145 KI210265 ENN79434.1 ERL95804.1 ERL96217.1 DQ910328 ABI83750.1 AAAB01008820 EAA05411.2 ATLV01011796 KE524723 KFB36136.1 KQ435821 KOX72351.1 GGFJ01009371 MBW58512.1 GFDF01003552 JAV10532.1 GANO01001175 JAB58696.1 AXCM01002011 KQ981512 KYN40837.1 GL440824 EFN65333.1 KQ976532 KYM81618.1 ADTU01026326 GL888284 EGI63302.1 CH477445 EAT40780.1 KQ979048 KYN23215.1 JXUM01059133 GAPW01004130 KQ562037 JAC09468.1 KXJ76844.1 GBGD01002844 JAC86045.1 LBMM01004764 KMQ92101.1 CM000160 EDW95638.1 KQ976780 KYN08400.1 CH933806 EDW15517.1 CP012526 ALC46538.1 UFQT01000413 UFQT01000418 SSX24075.1 SSX24140.1 CM000070 EAL28767.1 NNAY01000018 OXU31898.1 GDKW01000740 JAI55855.1 ACPB03020181 CH479185 EDW37930.1 GBXI01012588 JAD01704.1 GBYB01001514 JAG71281.1 GAMC01004456 JAC02100.1 KK107119 QOIP01000009 EZA58548.1 RLU18271.1 GDHF01001879 JAI50435.1 GAKP01009098 GAKP01009097 JAC49855.1 GFDF01003602 JAV10482.1 CH940650 EDW68073.1 GL446527 EFN87651.1 AE014297 AY058381 AAF52150.1 AAL13610.1 ALI30525.1 OUUW01000014 SPP88301.1 CM000364 EDX11580.1 CH480821 EDW54811.1 CH954181 EDV47760.1 GBHO01039472 GBHO01039471 GBHO01039469 GBRD01002433 GBRD01002432 JAG04132.1 JAG04133.1 JAG04135.1 JAG63388.1 CAQQ02141285 DS231872 EDS41417.1 CH916374 EDV91391.1 CH902617 EDV42855.1 GGFK01008833 MBW42154.1 GBBI01002449 JAC16263.1 GECL01002237 JAP03887.1 GDHF01028681 JAI23633.1 GFDL01008866 JAV26179.1 CH964232 EDW81571.1 GECU01010009 JAS97697.1 GFDL01010167 JAV24878.1 GECZ01025949 GECZ01014506 JAS43820.1 JAS55263.1 KQ434851 KZC08600.1 DS235806 EEB17313.1 CVRI01000070 CRL07221.1 JRES01000310 KNC32402.1 LRGB01000512 KZS18696.1 GDHC01014123 JAQ04506.1 CCAG010004727 GFWV01020286 MAA45014.1
ARD08865.1 SOQ34575.1 KQ459078 KPJ03821.1 AK401755 BAM18377.1 NWSH01000595 PCG75456.1 AGBW02014588 OWR41375.1 GGFJ01009199 MBW58340.1 GGFK01008052 MBW41373.1 GGFM01003760 MBW24511.1 GEZM01074367 JAV64583.1 KK852496 KDR22737.1 ADMH02002130 ETN58540.1 NEVH01020342 PNF21511.1 GAMD01002851 JAA98739.1 AXCN02000234 KQ971371 EFA10075.1 APGK01028591 KB740694 KI208145 KI210265 ENN79434.1 ERL95804.1 ERL96217.1 DQ910328 ABI83750.1 AAAB01008820 EAA05411.2 ATLV01011796 KE524723 KFB36136.1 KQ435821 KOX72351.1 GGFJ01009371 MBW58512.1 GFDF01003552 JAV10532.1 GANO01001175 JAB58696.1 AXCM01002011 KQ981512 KYN40837.1 GL440824 EFN65333.1 KQ976532 KYM81618.1 ADTU01026326 GL888284 EGI63302.1 CH477445 EAT40780.1 KQ979048 KYN23215.1 JXUM01059133 GAPW01004130 KQ562037 JAC09468.1 KXJ76844.1 GBGD01002844 JAC86045.1 LBMM01004764 KMQ92101.1 CM000160 EDW95638.1 KQ976780 KYN08400.1 CH933806 EDW15517.1 CP012526 ALC46538.1 UFQT01000413 UFQT01000418 SSX24075.1 SSX24140.1 CM000070 EAL28767.1 NNAY01000018 OXU31898.1 GDKW01000740 JAI55855.1 ACPB03020181 CH479185 EDW37930.1 GBXI01012588 JAD01704.1 GBYB01001514 JAG71281.1 GAMC01004456 JAC02100.1 KK107119 QOIP01000009 EZA58548.1 RLU18271.1 GDHF01001879 JAI50435.1 GAKP01009098 GAKP01009097 JAC49855.1 GFDF01003602 JAV10482.1 CH940650 EDW68073.1 GL446527 EFN87651.1 AE014297 AY058381 AAF52150.1 AAL13610.1 ALI30525.1 OUUW01000014 SPP88301.1 CM000364 EDX11580.1 CH480821 EDW54811.1 CH954181 EDV47760.1 GBHO01039472 GBHO01039471 GBHO01039469 GBRD01002433 GBRD01002432 JAG04132.1 JAG04133.1 JAG04135.1 JAG63388.1 CAQQ02141285 DS231872 EDS41417.1 CH916374 EDV91391.1 CH902617 EDV42855.1 GGFK01008833 MBW42154.1 GBBI01002449 JAC16263.1 GECL01002237 JAP03887.1 GDHF01028681 JAI23633.1 GFDL01008866 JAV26179.1 CH964232 EDW81571.1 GECU01010009 JAS97697.1 GFDL01010167 JAV24878.1 GECZ01025949 GECZ01014506 JAS43820.1 JAS55263.1 KQ434851 KZC08600.1 DS235806 EEB17313.1 CVRI01000070 CRL07221.1 JRES01000310 KNC32402.1 LRGB01000512 KZS18696.1 GDHC01014123 JAQ04506.1 CCAG010004727 GFWV01020286 MAA45014.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000027135
UP000192223
+ More
UP000000673 UP000069272 UP000075881 UP000075884 UP000075920 UP000235965 UP000075900 UP000075886 UP000007266 UP000019118 UP000030742 UP000076408 UP000075902 UP000076407 UP000007062 UP000075885 UP000075903 UP000030765 UP000053105 UP000075883 UP000005203 UP000078541 UP000000311 UP000078540 UP000005205 UP000007755 UP000008820 UP000078492 UP000069940 UP000249989 UP000036403 UP000002282 UP000078542 UP000009192 UP000092553 UP000001819 UP000215335 UP000015103 UP000008744 UP000075880 UP000053097 UP000279307 UP000002358 UP000008792 UP000008237 UP000000803 UP000268350 UP000192221 UP000000304 UP000001292 UP000008711 UP000015102 UP000002320 UP000001070 UP000007801 UP000007798 UP000095300 UP000076502 UP000009046 UP000095301 UP000183832 UP000037069 UP000076858 UP000092444 UP000092443
UP000000673 UP000069272 UP000075881 UP000075884 UP000075920 UP000235965 UP000075900 UP000075886 UP000007266 UP000019118 UP000030742 UP000076408 UP000075902 UP000076407 UP000007062 UP000075885 UP000075903 UP000030765 UP000053105 UP000075883 UP000005203 UP000078541 UP000000311 UP000078540 UP000005205 UP000007755 UP000008820 UP000078492 UP000069940 UP000249989 UP000036403 UP000002282 UP000078542 UP000009192 UP000092553 UP000001819 UP000215335 UP000015103 UP000008744 UP000075880 UP000053097 UP000279307 UP000002358 UP000008792 UP000008237 UP000000803 UP000268350 UP000192221 UP000000304 UP000001292 UP000008711 UP000015102 UP000002320 UP000001070 UP000007801 UP000007798 UP000095300 UP000076502 UP000009046 UP000095301 UP000183832 UP000037069 UP000076858 UP000092444 UP000092443
Pfam
PF03357 Snf7
Interpro
IPR005024
Snf7_fam
ProteinModelPortal
A0A1E1W5M1
A0A194RQL6
A0A1V0JHV4
A0A0N1PF08
I4DKD7
A0A2A4JUK8
+ More
A0A212EIR8 A0A2M4BZ49 A0A2M4AKR4 A0A2M3Z7N0 A0A1Y1KT70 A0A067RG49 A0A1W4WMF6 W5J6Q6 A0A182FL05 A0A182KFS0 A0A182NSJ0 A0A182VS48 A0A2J7PYU5 T1DG90 A0A182R892 A0A182QQS7 D6X057 N6TGA5 A0A182YLL9 Q06DJ7 A0A182TWV2 A0A182WW00 Q7QH42 A0A182P088 A0A182UQ78 A0A084VDU2 A0A0N1ITF9 A0A2M4BZQ8 A0A1L8DVS5 U5EX10 A0A182MPT5 A0A088AF24 A0A195FKQ0 E2AML3 A0A195BAQ3 A0A158NU97 F4WR85 Q171V6 A0A151JCE6 A0A023EK15 A0A069DQ93 A0A0J7KPB8 B4PTQ5 A0A151IQK9 B4K985 A0A0M4EP29 A0A336M1F5 Q295E5 A0A232FN59 A0A0P4VL65 T1IEU2 B4GM56 A0A182INF8 A0A0A1WRL9 A0A0C9QDW5 W8CBW2 A0A026WRE1 A0A0K8WHU3 A0A034W6U4 K7INY3 A0A1L8DVM1 B4LYX0 E2B9G8 Q9VN02 A0A3B0KSV6 A0A1W4VL71 B4QUZ9 B4I3L8 B3P1Y7 A0A0A9WH18 T1GBQ4 B0WAJ4 B4JV71 B3LXZ9 A0A2M4AMZ7 A0A023F592 A0A0V0G921 A0A0K8UAL6 A0A1Q3FF52 B4N924 A0A1B6JEX8 A0A1Q3FBK3 A0A1I8NV40 A0A1B6F0T6 A0A154PBN0 E0VVA7 A0A1I8MIU2 A0A1J1J485 A0A0L0CJM6 A0A162PB75 A0A146LBE1 A0A1B0FFU9 A0A1A9YQ12 A0A293MW15
A0A212EIR8 A0A2M4BZ49 A0A2M4AKR4 A0A2M3Z7N0 A0A1Y1KT70 A0A067RG49 A0A1W4WMF6 W5J6Q6 A0A182FL05 A0A182KFS0 A0A182NSJ0 A0A182VS48 A0A2J7PYU5 T1DG90 A0A182R892 A0A182QQS7 D6X057 N6TGA5 A0A182YLL9 Q06DJ7 A0A182TWV2 A0A182WW00 Q7QH42 A0A182P088 A0A182UQ78 A0A084VDU2 A0A0N1ITF9 A0A2M4BZQ8 A0A1L8DVS5 U5EX10 A0A182MPT5 A0A088AF24 A0A195FKQ0 E2AML3 A0A195BAQ3 A0A158NU97 F4WR85 Q171V6 A0A151JCE6 A0A023EK15 A0A069DQ93 A0A0J7KPB8 B4PTQ5 A0A151IQK9 B4K985 A0A0M4EP29 A0A336M1F5 Q295E5 A0A232FN59 A0A0P4VL65 T1IEU2 B4GM56 A0A182INF8 A0A0A1WRL9 A0A0C9QDW5 W8CBW2 A0A026WRE1 A0A0K8WHU3 A0A034W6U4 K7INY3 A0A1L8DVM1 B4LYX0 E2B9G8 Q9VN02 A0A3B0KSV6 A0A1W4VL71 B4QUZ9 B4I3L8 B3P1Y7 A0A0A9WH18 T1GBQ4 B0WAJ4 B4JV71 B3LXZ9 A0A2M4AMZ7 A0A023F592 A0A0V0G921 A0A0K8UAL6 A0A1Q3FF52 B4N924 A0A1B6JEX8 A0A1Q3FBK3 A0A1I8NV40 A0A1B6F0T6 A0A154PBN0 E0VVA7 A0A1I8MIU2 A0A1J1J485 A0A0L0CJM6 A0A162PB75 A0A146LBE1 A0A1B0FFU9 A0A1A9YQ12 A0A293MW15
PDB
3FRT
E-value=4.23198e-45,
Score=454
Ontologies
GO
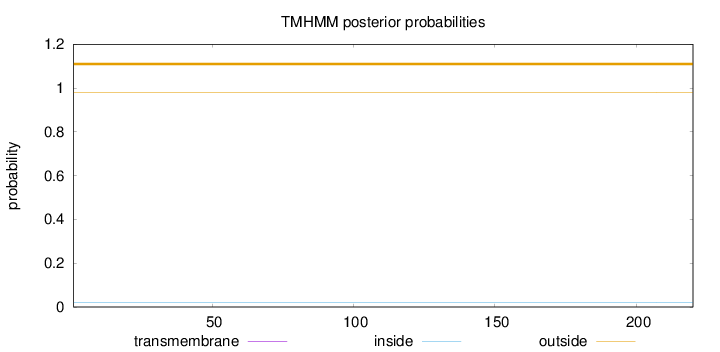
Topology
Length:
220
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01929
outside
1 - 220
Population Genetic Test Statistics
Pi
236.809492
Theta
162.429738
Tajima's D
1.507206
CLR
0.082212
CSRT
0.789010549472526
Interpretation
Uncertain
