Gene
KWMTBOMO15587
Pre Gene Modal
BGIBMGA010828
Annotation
PREDICTED:_xenotropic_and_polytropic_retrovirus_receptor_1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.92
Sequence
CDS
ATGAAGAAACTAAGAGTGCCACCTCTCGGGGAGCAGCAGAGTCCGTGGACCACATTCAAAGTTGGATTGTTTTCAGGGTCATTTATAGTATTAATCATCTCAGTTGTTTTGTCAGCTCTATTTCACGAAGGTGGATCGAGCGATATCAAAACAGCCATTCGATTATACAGAGGTCCGTTCCTCTTGGTCGAGTTCATATTTTTGATAGGCATTAACGTATACGGATGGCGGTCGTCAGGAGTGAACCACGTGCTAATATTCGAACTAGACCCAAGGAATCACTTGTCCGAGCAGCATTTAATGGAATTAGCGGCAATTTTCGGTGTCGTATGGGCGCTTAGTATATTAAGTTTTATATACAGCGCGAGCCTTAGCATACCGCCTTTTGTGAATCCCCTAGCGCTGGTTGTGATAATGCTTGCCTTCTTGATGAACCCATTCAAGGTTTTCCGGCATGAAGCCAGATTCTGGTTTTTGAGGATATGTGGTCGGATCCTCGCAGCGCCATTCCTGCCCGTGCTATTCGCCGATTTCTGGCTGGCCGACCAATGGAATTCGTTCACGGCCGCCTTCGTAGACTTTCACTATCTGATCTCTTTCTACGTGGCGGGCGGTGATTGGTTCAAAGTTAATAACGAGTTCGAAGCGGCCCAATGGTTCGTGTTGAGCCGCGCGGTCGTGAGCGTGGTCCCGGCCTGGACCCGGTTCTGGCAGTGTCTGCGCCGGTACCGGGACAGCCGGGAGGCGTTCCCGCATCTCGTCAACGCCGGGAAGTACTCGACCACGTTTTTCGTGGTTCTATTCGCTACGTTGAAGACTTTGAACAATCATAAATATCCAGATTCGTCTTTCGACAATCCGTTTCTGTACGCGTGGTTCGCGTGTCAGCTCGTGTCGTCCGTGTACACGTACACGTGGGACGTGAAAATGGACTGGGGTCTGTTCAGCTGTGGGCCCAGCGCTGAGAATAAGTTCCTCAGGGAGGAGATCGTCTACAGTCCGGGGTCTTACTACTTTGCGATAGTAGAGGATTTCGTATTACGTTTCATATGGACTGTTTCGTTCGTTCTCACGGAGAATAAATATGTCGGCTCCGAAACTATGACCTCCATATTGGCTCCCCTGGAAGTTTTCAGACGTTTCATCTGGAATTTCTTCCGTCTCGAGAACGAGCACCTGAACAACTGCGGCAAGTTCCGCGCCGTCCGGGACATCTCCGTGGCGCCCCTGGATTCTTCCGACCAGGCCGACATCATCCGGATGATGGACCATCCCGATGGAGTTATAAACAGATTAACGAAGAAGAAAAACGCAAAAAAGACAAAGGAAATCGACCGTCGGCCGCTGCTGAAGAAGGAATCGATACAAGATCTTCAATTAGAACTCGACTTGTCCTCCAAAATGATGTAA
Protein
MKKLRVPPLGEQQSPWTTFKVGLFSGSFIVLIISVVLSALFHEGGSSDIKTAIRLYRGPFLLVEFIFLIGINVYGWRSSGVNHVLIFELDPRNHLSEQHLMELAAIFGVVWALSILSFIYSASLSIPPFVNPLALVVIMLAFLMNPFKVFRHEARFWFLRICGRILAAPFLPVLFADFWLADQWNSFTAAFVDFHYLISFYVAGGDWFKVNNEFEAAQWFVLSRAVVSVVPAWTRFWQCLRRYRDSREAFPHLVNAGKYSTTFFVVLFATLKTLNNHKYPDSSFDNPFLYAWFACQLVSSVYTYTWDVKMDWGLFSCGPSAENKFLREEIVYSPGSYYFAIVEDFVLRFIWTVSFVLTENKYVGSETMTSILAPLEVFRRFIWNFFRLENEHLNNCGKFRAVRDISVAPLDSSDQADIIRMMDHPDGVINRLTKKKNAKKTKEIDRRPLLKKESIQDLQLELDLSSKMM
Summary
Uniprot
H9JMS5
A0A1E1WMX8
A0A2H1V2S5
A0A2A4JTU4
A0A212EIQ9
A0A194RU09
+ More
A0A1Y1L9U8 B0WSZ6 A0A2J7QTZ5 A0A1Q3F8B0 A0A1Q3F8F7 A0A182GIC7 Q16QS0 A0A2J7QTY5 A0A0A1X828 A0A0K8TWG6 A0A182HDB8 A0A2J7QTZ3 A0A067QH79 A0A182J1K0 A0A084WFL5 A0A182MV72 A0A182RKP9 A0A182N0N0 A0A182QY00 A0A182YH08 A0A182T7I1 A0A0C9QUC2 A0A182VD08 Q7QEH3 A0A1W4XHM7 T1PAF2 A0A1I8Q6F8 B4JJ94 A0A182KED6 B4NCH8 A0A1B6CLC4 A0A336MAQ2 B4L299 A0A1A9YKA6 A0A0M5JE02 A0A0J7L795 W8BL46 B3NTA1 A0A0N1IAA6 A0A1B6GBR3 B4M6R4 D7EJG4 B4Q2N3 A0A1B6IVX8 A0A158NFD2 A0A1A9W6A3 A0A1W4UNM9 F4WUI9 A0A1B0FG65 A0A3B0J8A2 A0A2A3E3H5 A0A087ZYJ9 K7IW66 Q29H73 A0A1A9V6M3 A0A1B0AXR1 N6U276 A0A1J1IXE6 A0A3L8DGJ9 E2A6B8 A0A1B0A5H5 A0A232FDK4 A0A3B0JD69 A0A2M4BG97 A0A2M4AJ02 A0A067QY66 Q9VWZ2 A0A1B0CW89 A0A2M3ZH92 A0A2J7PCB6 A0A182FJX0 W5J9P8 A0A1B6J659 A0A023F4Q6 A0A182W5W8 A0A1Y1L9W9 A0A1L8DUN1 A0A1L8DUM5 A0A1B6D1A0 A0A1B6CDV4 A0A1D2N4I0 A0A226ELL4 T1HVP6 E0W1Q8 T1JMQ6 A0A023F535 T1I243 D6WFX6 A0A182Q5C0 A0A084WKL4 A0A1W4X7G3 A0A182JZV7 A0A158NT11 A0A0K8VFF5
A0A1Y1L9U8 B0WSZ6 A0A2J7QTZ5 A0A1Q3F8B0 A0A1Q3F8F7 A0A182GIC7 Q16QS0 A0A2J7QTY5 A0A0A1X828 A0A0K8TWG6 A0A182HDB8 A0A2J7QTZ3 A0A067QH79 A0A182J1K0 A0A084WFL5 A0A182MV72 A0A182RKP9 A0A182N0N0 A0A182QY00 A0A182YH08 A0A182T7I1 A0A0C9QUC2 A0A182VD08 Q7QEH3 A0A1W4XHM7 T1PAF2 A0A1I8Q6F8 B4JJ94 A0A182KED6 B4NCH8 A0A1B6CLC4 A0A336MAQ2 B4L299 A0A1A9YKA6 A0A0M5JE02 A0A0J7L795 W8BL46 B3NTA1 A0A0N1IAA6 A0A1B6GBR3 B4M6R4 D7EJG4 B4Q2N3 A0A1B6IVX8 A0A158NFD2 A0A1A9W6A3 A0A1W4UNM9 F4WUI9 A0A1B0FG65 A0A3B0J8A2 A0A2A3E3H5 A0A087ZYJ9 K7IW66 Q29H73 A0A1A9V6M3 A0A1B0AXR1 N6U276 A0A1J1IXE6 A0A3L8DGJ9 E2A6B8 A0A1B0A5H5 A0A232FDK4 A0A3B0JD69 A0A2M4BG97 A0A2M4AJ02 A0A067QY66 Q9VWZ2 A0A1B0CW89 A0A2M3ZH92 A0A2J7PCB6 A0A182FJX0 W5J9P8 A0A1B6J659 A0A023F4Q6 A0A182W5W8 A0A1Y1L9W9 A0A1L8DUN1 A0A1L8DUM5 A0A1B6D1A0 A0A1B6CDV4 A0A1D2N4I0 A0A226ELL4 T1HVP6 E0W1Q8 T1JMQ6 A0A023F535 T1I243 D6WFX6 A0A182Q5C0 A0A084WKL4 A0A1W4X7G3 A0A182JZV7 A0A158NT11 A0A0K8VFF5
Pubmed
19121390
22118469
26354079
28004739
26483478
17510324
+ More
25830018 24845553 24438588 25244985 12364791 14747013 17210077 25315136 17994087 18057021 24495485 18362917 19820115 17550304 21347285 21719571 20075255 15632085 23185243 23537049 30249741 20798317 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25474469 27289101 20566863
25830018 24845553 24438588 25244985 12364791 14747013 17210077 25315136 17994087 18057021 24495485 18362917 19820115 17550304 21347285 21719571 20075255 15632085 23185243 23537049 30249741 20798317 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25474469 27289101 20566863
EMBL
BABH01016797
BABH01016798
GDQN01002700
JAT88354.1
ODYU01000194
SOQ34574.1
+ More
NWSH01000595 PCG75455.1 AGBW02014588 OWR41376.1 KQ459875 KPJ19606.1 GEZM01061507 GEZM01061506 JAV70423.1 DS232078 EDS34129.1 NEVH01011192 PNF32043.1 GFDL01011269 JAV23776.1 GFDL01011228 JAV23817.1 JXUM01065729 JXUM01065730 JXUM01065731 JXUM01065732 JXUM01065733 JXUM01065734 JXUM01065735 JXUM01065736 CH477736 EAT36732.1 PNF32047.1 GBXI01006823 GBXI01006487 JAD07469.1 JAD07805.1 GDHF01033515 GDHF01024258 JAI18799.1 JAI28056.1 JXUM01034121 JXUM01034122 JXUM01034123 KQ561012 KXJ80041.1 PNF32044.1 KK853424 KDR07686.1 ATLV01023372 KE525342 KFB49009.1 AXCM01000747 AXCN02002234 GBYB01007349 GBYB01008920 JAG77116.1 JAG78687.1 AAAB01008847 EAA06862.3 KA645674 AFP60303.1 CH916370 EDV99646.1 CH964239 EDW82537.1 GEDC01023048 JAS14250.1 UFQT01000315 SSX23118.1 CH933810 EDW07760.1 KRF94197.1 KRF94198.1 KRF94199.1 CP012528 ALC49868.1 LBMM01000318 KMR01389.1 GAMC01016196 JAB90359.1 CH954180 EDV46552.1 KQS30032.1 KQ459078 KPJ03822.1 GECZ01009890 JAS59879.1 CH940653 EDW62481.1 KRF80678.1 DS497704 EFA12730.1 CM000162 EDX02674.1 KRK06911.1 GECU01016706 JAS91000.1 ADTU01014274 GL888365 EGI62154.1 CCAG010014756 OUUW01000003 SPP78544.1 KZ288400 PBC26303.1 CH379064 EAL31885.1 KRT06642.1 KRT06643.1 KRT06644.1 JXJN01005341 APGK01042726 KB741007 KB630653 KB631811 ENN75640.1 ERL83766.1 ERL86361.1 CVRI01000063 CRL04244.1 QOIP01000009 RLU19039.1 GL437123 EFN71003.1 NNAY01000414 OXU28573.1 SPP78543.1 GGFJ01002921 MBW52062.1 GGFK01007430 MBW40751.1 KK853117 KDR11153.1 AE014298 AY051791 AAF48793.2 AAK93215.1 AJWK01032013 AJWK01032014 AJWK01032015 AJWK01032016 AJWK01032017 AJWK01032018 AJWK01032019 GGFM01007128 MBW27879.1 NEVH01027063 PNF13968.1 ADMH02001869 ETN60726.1 GECU01013048 JAS94658.1 GBBI01002734 JAC15978.1 GEZM01061505 JAV70424.1 GFDF01003943 JAV10141.1 GFDF01003942 JAV10142.1 GEDC01017844 JAS19454.1 GEDC01025666 JAS11632.1 LJIJ01000228 ODN00179.1 LNIX01000003 OXA58108.1 ACPB03010092 DS235873 EEB19640.1 JH431929 GBBI01002553 JAC16159.1 ACPB03000284 KQ971328 EEZ99778.1 AXCN02000985 ATLV01024126 KE525349 KFB50758.1 ADTU01025470 GDHF01014630 JAI37684.1
NWSH01000595 PCG75455.1 AGBW02014588 OWR41376.1 KQ459875 KPJ19606.1 GEZM01061507 GEZM01061506 JAV70423.1 DS232078 EDS34129.1 NEVH01011192 PNF32043.1 GFDL01011269 JAV23776.1 GFDL01011228 JAV23817.1 JXUM01065729 JXUM01065730 JXUM01065731 JXUM01065732 JXUM01065733 JXUM01065734 JXUM01065735 JXUM01065736 CH477736 EAT36732.1 PNF32047.1 GBXI01006823 GBXI01006487 JAD07469.1 JAD07805.1 GDHF01033515 GDHF01024258 JAI18799.1 JAI28056.1 JXUM01034121 JXUM01034122 JXUM01034123 KQ561012 KXJ80041.1 PNF32044.1 KK853424 KDR07686.1 ATLV01023372 KE525342 KFB49009.1 AXCM01000747 AXCN02002234 GBYB01007349 GBYB01008920 JAG77116.1 JAG78687.1 AAAB01008847 EAA06862.3 KA645674 AFP60303.1 CH916370 EDV99646.1 CH964239 EDW82537.1 GEDC01023048 JAS14250.1 UFQT01000315 SSX23118.1 CH933810 EDW07760.1 KRF94197.1 KRF94198.1 KRF94199.1 CP012528 ALC49868.1 LBMM01000318 KMR01389.1 GAMC01016196 JAB90359.1 CH954180 EDV46552.1 KQS30032.1 KQ459078 KPJ03822.1 GECZ01009890 JAS59879.1 CH940653 EDW62481.1 KRF80678.1 DS497704 EFA12730.1 CM000162 EDX02674.1 KRK06911.1 GECU01016706 JAS91000.1 ADTU01014274 GL888365 EGI62154.1 CCAG010014756 OUUW01000003 SPP78544.1 KZ288400 PBC26303.1 CH379064 EAL31885.1 KRT06642.1 KRT06643.1 KRT06644.1 JXJN01005341 APGK01042726 KB741007 KB630653 KB631811 ENN75640.1 ERL83766.1 ERL86361.1 CVRI01000063 CRL04244.1 QOIP01000009 RLU19039.1 GL437123 EFN71003.1 NNAY01000414 OXU28573.1 SPP78543.1 GGFJ01002921 MBW52062.1 GGFK01007430 MBW40751.1 KK853117 KDR11153.1 AE014298 AY051791 AAF48793.2 AAK93215.1 AJWK01032013 AJWK01032014 AJWK01032015 AJWK01032016 AJWK01032017 AJWK01032018 AJWK01032019 GGFM01007128 MBW27879.1 NEVH01027063 PNF13968.1 ADMH02001869 ETN60726.1 GECU01013048 JAS94658.1 GBBI01002734 JAC15978.1 GEZM01061505 JAV70424.1 GFDF01003943 JAV10141.1 GFDF01003942 JAV10142.1 GEDC01017844 JAS19454.1 GEDC01025666 JAS11632.1 LJIJ01000228 ODN00179.1 LNIX01000003 OXA58108.1 ACPB03010092 DS235873 EEB19640.1 JH431929 GBBI01002553 JAC16159.1 ACPB03000284 KQ971328 EEZ99778.1 AXCN02000985 ATLV01024126 KE525349 KFB50758.1 ADTU01025470 GDHF01014630 JAI37684.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000002320
UP000235965
+ More
UP000069940 UP000008820 UP000249989 UP000027135 UP000075880 UP000030765 UP000075883 UP000075900 UP000075884 UP000075886 UP000076408 UP000075901 UP000075903 UP000007062 UP000192223 UP000095301 UP000095300 UP000001070 UP000075881 UP000007798 UP000009192 UP000092443 UP000092553 UP000036403 UP000008711 UP000053268 UP000008792 UP000007266 UP000002282 UP000005205 UP000091820 UP000192221 UP000007755 UP000092444 UP000268350 UP000242457 UP000005203 UP000002358 UP000001819 UP000078200 UP000092460 UP000019118 UP000030742 UP000183832 UP000279307 UP000000311 UP000092445 UP000215335 UP000000803 UP000092461 UP000069272 UP000000673 UP000075920 UP000094527 UP000198287 UP000015103 UP000009046
UP000069940 UP000008820 UP000249989 UP000027135 UP000075880 UP000030765 UP000075883 UP000075900 UP000075884 UP000075886 UP000076408 UP000075901 UP000075903 UP000007062 UP000192223 UP000095301 UP000095300 UP000001070 UP000075881 UP000007798 UP000009192 UP000092443 UP000092553 UP000036403 UP000008711 UP000053268 UP000008792 UP000007266 UP000002282 UP000005205 UP000091820 UP000192221 UP000007755 UP000092444 UP000268350 UP000242457 UP000005203 UP000002358 UP000001819 UP000078200 UP000092460 UP000019118 UP000030742 UP000183832 UP000279307 UP000000311 UP000092445 UP000215335 UP000000803 UP000092461 UP000069272 UP000000673 UP000075920 UP000094527 UP000198287 UP000015103 UP000009046
Pfam
Interpro
IPR004342
EXS_C
+ More
IPR004331 SPX_dom
IPR036388 WH-like_DNA-bd_sf
IPR035810 PEBP_euk
IPR036610 PEBP-like_sf
IPR008253 Marvel
IPR036390 WH_DNA-bd_sf
IPR035979 RBD_domain_sf
IPR006630 La_HTH
IPR002344 Lupus_La
IPR008914 PEBP
IPR034910 LARP7_RRM2
IPR014886 La_RRM
IPR029060 PIN-like_dom_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR002716 PIN_dom
IPR004331 SPX_dom
IPR036388 WH-like_DNA-bd_sf
IPR035810 PEBP_euk
IPR036610 PEBP-like_sf
IPR008253 Marvel
IPR036390 WH_DNA-bd_sf
IPR035979 RBD_domain_sf
IPR006630 La_HTH
IPR002344 Lupus_La
IPR008914 PEBP
IPR034910 LARP7_RRM2
IPR014886 La_RRM
IPR029060 PIN-like_dom_sf
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR002716 PIN_dom
Gene 3D
ProteinModelPortal
H9JMS5
A0A1E1WMX8
A0A2H1V2S5
A0A2A4JTU4
A0A212EIQ9
A0A194RU09
+ More
A0A1Y1L9U8 B0WSZ6 A0A2J7QTZ5 A0A1Q3F8B0 A0A1Q3F8F7 A0A182GIC7 Q16QS0 A0A2J7QTY5 A0A0A1X828 A0A0K8TWG6 A0A182HDB8 A0A2J7QTZ3 A0A067QH79 A0A182J1K0 A0A084WFL5 A0A182MV72 A0A182RKP9 A0A182N0N0 A0A182QY00 A0A182YH08 A0A182T7I1 A0A0C9QUC2 A0A182VD08 Q7QEH3 A0A1W4XHM7 T1PAF2 A0A1I8Q6F8 B4JJ94 A0A182KED6 B4NCH8 A0A1B6CLC4 A0A336MAQ2 B4L299 A0A1A9YKA6 A0A0M5JE02 A0A0J7L795 W8BL46 B3NTA1 A0A0N1IAA6 A0A1B6GBR3 B4M6R4 D7EJG4 B4Q2N3 A0A1B6IVX8 A0A158NFD2 A0A1A9W6A3 A0A1W4UNM9 F4WUI9 A0A1B0FG65 A0A3B0J8A2 A0A2A3E3H5 A0A087ZYJ9 K7IW66 Q29H73 A0A1A9V6M3 A0A1B0AXR1 N6U276 A0A1J1IXE6 A0A3L8DGJ9 E2A6B8 A0A1B0A5H5 A0A232FDK4 A0A3B0JD69 A0A2M4BG97 A0A2M4AJ02 A0A067QY66 Q9VWZ2 A0A1B0CW89 A0A2M3ZH92 A0A2J7PCB6 A0A182FJX0 W5J9P8 A0A1B6J659 A0A023F4Q6 A0A182W5W8 A0A1Y1L9W9 A0A1L8DUN1 A0A1L8DUM5 A0A1B6D1A0 A0A1B6CDV4 A0A1D2N4I0 A0A226ELL4 T1HVP6 E0W1Q8 T1JMQ6 A0A023F535 T1I243 D6WFX6 A0A182Q5C0 A0A084WKL4 A0A1W4X7G3 A0A182JZV7 A0A158NT11 A0A0K8VFF5
A0A1Y1L9U8 B0WSZ6 A0A2J7QTZ5 A0A1Q3F8B0 A0A1Q3F8F7 A0A182GIC7 Q16QS0 A0A2J7QTY5 A0A0A1X828 A0A0K8TWG6 A0A182HDB8 A0A2J7QTZ3 A0A067QH79 A0A182J1K0 A0A084WFL5 A0A182MV72 A0A182RKP9 A0A182N0N0 A0A182QY00 A0A182YH08 A0A182T7I1 A0A0C9QUC2 A0A182VD08 Q7QEH3 A0A1W4XHM7 T1PAF2 A0A1I8Q6F8 B4JJ94 A0A182KED6 B4NCH8 A0A1B6CLC4 A0A336MAQ2 B4L299 A0A1A9YKA6 A0A0M5JE02 A0A0J7L795 W8BL46 B3NTA1 A0A0N1IAA6 A0A1B6GBR3 B4M6R4 D7EJG4 B4Q2N3 A0A1B6IVX8 A0A158NFD2 A0A1A9W6A3 A0A1W4UNM9 F4WUI9 A0A1B0FG65 A0A3B0J8A2 A0A2A3E3H5 A0A087ZYJ9 K7IW66 Q29H73 A0A1A9V6M3 A0A1B0AXR1 N6U276 A0A1J1IXE6 A0A3L8DGJ9 E2A6B8 A0A1B0A5H5 A0A232FDK4 A0A3B0JD69 A0A2M4BG97 A0A2M4AJ02 A0A067QY66 Q9VWZ2 A0A1B0CW89 A0A2M3ZH92 A0A2J7PCB6 A0A182FJX0 W5J9P8 A0A1B6J659 A0A023F4Q6 A0A182W5W8 A0A1Y1L9W9 A0A1L8DUN1 A0A1L8DUM5 A0A1B6D1A0 A0A1B6CDV4 A0A1D2N4I0 A0A226ELL4 T1HVP6 E0W1Q8 T1JMQ6 A0A023F535 T1I243 D6WFX6 A0A182Q5C0 A0A084WKL4 A0A1W4X7G3 A0A182JZV7 A0A158NT11 A0A0K8VFF5
Ontologies
GO
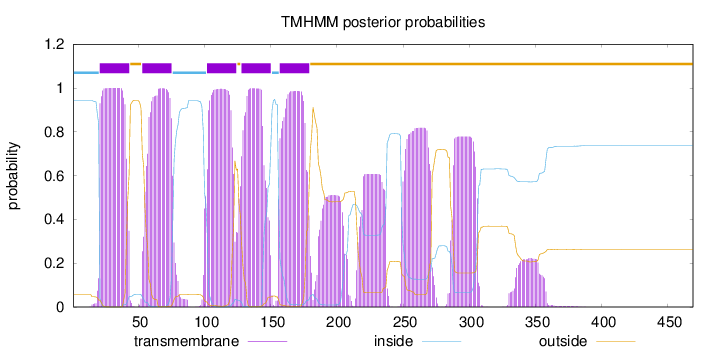
Topology
Length:
469
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
166.91952
Exp number, first 60 AAs:
26.74411
Total prob of N-in:
0.94300
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 52
TMhelix
53 - 75
inside
76 - 101
TMhelix
102 - 124
outside
125 - 127
TMhelix
128 - 150
inside
151 - 156
TMhelix
157 - 179
outside
180 - 469
Population Genetic Test Statistics
Pi
231.820395
Theta
176.17245
Tajima's D
1.208457
CLR
0.478395
CSRT
0.713514324283786
Interpretation
Uncertain
