Gene
KWMTBOMO15582
Pre Gene Modal
BGIBMGA010826
Annotation
PREDICTED:_ABC_transporter_G_family_member_23-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.543
Sequence
CDS
ATGGCGGTCCGAGAAGCAGCAGTTATCGTAGAGTGTGCCAACAAGTCATATGGACAACAGAACGTGCTCAACGACCTAGACTTGACCGTCGAAAAGGGCACTATATATGGGCTGCTTGGTCCGTCGGGCTGTGGGAAAACGACGTTGCTCTCCTGCATAGTCGGAAGACGGAGCCTTGATAGTGGCAATGTCTGGGTGCTTGGTGGGAAGCCAGGAGAGAAGGGCAGCGGTGTGCCTGGCCCCAGAGTGGGATATATGCCGCAGGACATAGCTTTAGTTGGAGAGTTCACGGTCAGAGATGCTGTATACTACTTCGGTCGCATATACGGCATGGAGCATAAGAAGATGACGGAGCGCTTTGAGTTCCTCAGCTCGTTACTGGACCTGCCCAGTGGGAATCGACTCATTAAAAATTGCTCCGGAGGGCAGCAACGAAGAGTCTCGTTGGCAGCTGCTTTAGTCCACGACCCAGAGCTGTTAATCTTAGACGAACCGACGGTTGGATTGGACCCTGTATTGAGGGAGCAAATCTGGAAGTTCCTGACTGATGTCGCACAGCAGGGGACCACGGTTATCATCACAACGCACTATATCGATGAAACAAAACAAGCGCACAAGATAGGCTTACTGCGAGATGGACAGTTGCTGGCGGAAGATTCTCCAGATGAAATTCTGAGAAAGTTCGAGTGCAATACCCTTGAGGAGGCCTTTCTTAAGCTTGCCTTAGCTCAATATGAAATTCCACAACGAAGACCCCCACCGACAGTGTCTCCGGACGTGATCCCGGAAAGCAGACAGATCGAGAGCGTGTACGAGTCAAGGGAAACCTTCAATGCCGTGAACGGGAGCACTGATGTTTTAACAAAGAAAGAAAAGTCGATAAGGAGCAGAAATTTGTCAAGGTCCAGATACAAAGCCGTATTTATTAAGTGCATACAACAATTCTGTAGACATCCTGGGGGCCTAATATTCTCTATACTGTTTCCTATAATCGAGGTGGTCGCGTTCTTCCTGGCGGTGGGACACGACCCGAGGGACCTCCAGATCGCAGTCGTCAATGAAGAGGCGGTCTTCTCGCCGTTTGGCTTAGATATCTGTAGGAGAAGTGACTTACAAACGACAAAGTTGTTAAAGGACGGCACGTGTGAACTGTACATGCTGGGCTGCTGGTTCCTAGAAGAAATGGAGAACAAGGAATTGGTTCAGACGGTGTATAACAGTACTGAGGAGGCGAAAAGCGCCGTGGCAACTGGCAAGCTCTACGCCGCCCTCCACTTCGCTCAGAACTTCAGTGAGGCACTCGGGTTGCGGGTCGCTGAGGGAGAAGTCGCGGACCGCACGCTCGCCGAGAGCAACATCAGGGTCTGGCTCGACATGACCAATCACCAAATATCATATTTCATAAAGCAGCAGCTACACAAAGCTTACGAGAGCTTCACAAAGCGCGTGATGAGAGCTTGCGACAGACCCGAAAACATCATACAGATTCCGGTCAACTTCCACGATCCGATCTACGGTAAGAAGGAGCCGGAAATGGTTGGATACATGGCTCCCGGCATCTTGATCACGATAATATTCTTCCTGCCGAGCGTGGTGACGTCGACGCTGATGATCGGCGACCGGCTGGAGGGCGTGTGGGAGCGCAGCGCCGTGGCGGGCGTGCGGCCGCGCGAGATGCTCAACGTGCACATCGCGCTGCAGTCCGTCGTCATAGTGGTGCAGACAATCGAGATGATGGTGTTAGCGTTCGCTGGTTACGGTATACCGTCTAGGGGTTCCAATATCACGTGCGGGATCCTCCTGTTCCTCCAGGGTCTGTCGGGGATGTGCTACGGGTTCCTTCTGTCAGTCTACTGCACCAGTCACACGATGGCGTTCTTCGTTGCCACTGGAAGTTTCTATCCTATGATACTACTCTGCGGAATATTATGGCCCCTAGAAGGCATGTCTGGGGGGCTACGATACTTCGCGCTGGCCCTACCATTTACGATACCATCGAATTCCCTTCGAGATATGTTCGAGAAGGGAGCTTCTATCACCGACTCTACAGTTTATAACGGCTTCTTAGTCACATTTCTGTGGATATTCATCACTTTAGGCTTGATATTGTTACGATTGAAATATAAGAAAACTTAA
Protein
MAVREAAVIVECANKSYGQQNVLNDLDLTVEKGTIYGLLGPSGCGKTTLLSCIVGRRSLDSGNVWVLGGKPGEKGSGVPGPRVGYMPQDIALVGEFTVRDAVYYFGRIYGMEHKKMTERFEFLSSLLDLPSGNRLIKNCSGGQQRRVSLAAALVHDPELLILDEPTVGLDPVLREQIWKFLTDVAQQGTTVIITTHYIDETKQAHKIGLLRDGQLLAEDSPDEILRKFECNTLEEAFLKLALAQYEIPQRRPPPTVSPDVIPESRQIESVYESRETFNAVNGSTDVLTKKEKSIRSRNLSRSRYKAVFIKCIQQFCRHPGGLIFSILFPIIEVVAFFLAVGHDPRDLQIAVVNEEAVFSPFGLDICRRSDLQTTKLLKDGTCELYMLGCWFLEEMENKELVQTVYNSTEEAKSAVATGKLYAALHFAQNFSEALGLRVAEGEVADRTLAESNIRVWLDMTNHQISYFIKQQLHKAYESFTKRVMRACDRPENIIQIPVNFHDPIYGKKEPEMVGYMAPGILITIIFFLPSVVTSTLMIGDRLEGVWERSAVAGVRPREMLNVHIALQSVVIVVQTIEMMVLAFAGYGIPSRGSNITCGILLFLQGLSGMCYGFLLSVYCTSHTMAFFVATGSFYPMILLCGILWPLEGMSGGLRYFALALPFTIPSNSLRDMFEKGASITDSTVYNGFLVTFLWIFITLGLILLRLKYKKT
Summary
Uniprot
A0A0N1PG95
A0A212F4L4
A0A2A4JSP6
A0A194RQ20
A0A1L8DEW7
A0A1L8DF67
+ More
A0A1B0D2N6 A0A195CTG2 A0A182JPM5 A0A182P559 A0A182X0C0 A0A158NZH0 A0A182U3I1 A0A182R677 A0A182YAQ4 A0A2M3Z376 A0A182QE61 A0A182N442 A0A1J1IM12 A0A151WEU3 A0A1B0CGT5 A0A0K8TRC9 A0A182J8X7 Q7PYQ4 A0A067R2A0 A0A182FSU0 A0A182LV52 F4WLA0 A0A182VZ62 K7INT0 A0A1W4WF93 A0A139WBS7 W5J5L5 A0A139WBU4 A0A232FC36 A0A0L7QYQ1 K7J6H5 A0A1I8QDG6 A0A0U9HSE9 W8BBX8 A0A0K8VH42 A0A0A1XF64 A0A034WIK2 A0A0L0BZA0 A0A084VG16 A0A1B6DBU3 T1P8Q9 Q16GC7 D6X0P2 W8B731 W8AIT3 A0A034VCZ0 A0A3R5VSJ0 A0A0K8UCG2 A0A195CRZ4 A0A0L0CHB5 A0A1A9X055 A0A2H8TU48 A0A0G3FE97 A0A0A1XFV5 A0A1A9YT18 A0A0A1XDD6 E2BHE9 A0A1B0B3C6 J9K0F0 A0A1A9ZZ65 E0VTJ3 A0A2W1BZQ6 A0A151IYL4 A0A2A4JA19 A0A1I8PDS1 T1PDS1 A0A0N1INK1 A0A2H4TEV3 A0A1A9VFR8 V9IML8 E0VTI8 A0A0C9R7B6 A0A1Q3G4A7 A0A195CRT1 B4MV32 A0A2A3E0L0 A0A0M8ZZM1 F4WL98 A0A3B0JVB2 A0A154PSW7 E9J737 F4WL95 A0A151WUX0 A0A151JF56 F4WL93 A0A1B0A3U4 B3MML8 E2BHE8 A0A026WE32 A0A310SQQ1 B4JC64
A0A1B0D2N6 A0A195CTG2 A0A182JPM5 A0A182P559 A0A182X0C0 A0A158NZH0 A0A182U3I1 A0A182R677 A0A182YAQ4 A0A2M3Z376 A0A182QE61 A0A182N442 A0A1J1IM12 A0A151WEU3 A0A1B0CGT5 A0A0K8TRC9 A0A182J8X7 Q7PYQ4 A0A067R2A0 A0A182FSU0 A0A182LV52 F4WLA0 A0A182VZ62 K7INT0 A0A1W4WF93 A0A139WBS7 W5J5L5 A0A139WBU4 A0A232FC36 A0A0L7QYQ1 K7J6H5 A0A1I8QDG6 A0A0U9HSE9 W8BBX8 A0A0K8VH42 A0A0A1XF64 A0A034WIK2 A0A0L0BZA0 A0A084VG16 A0A1B6DBU3 T1P8Q9 Q16GC7 D6X0P2 W8B731 W8AIT3 A0A034VCZ0 A0A3R5VSJ0 A0A0K8UCG2 A0A195CRZ4 A0A0L0CHB5 A0A1A9X055 A0A2H8TU48 A0A0G3FE97 A0A0A1XFV5 A0A1A9YT18 A0A0A1XDD6 E2BHE9 A0A1B0B3C6 J9K0F0 A0A1A9ZZ65 E0VTJ3 A0A2W1BZQ6 A0A151IYL4 A0A2A4JA19 A0A1I8PDS1 T1PDS1 A0A0N1INK1 A0A2H4TEV3 A0A1A9VFR8 V9IML8 E0VTI8 A0A0C9R7B6 A0A1Q3G4A7 A0A195CRT1 B4MV32 A0A2A3E0L0 A0A0M8ZZM1 F4WL98 A0A3B0JVB2 A0A154PSW7 E9J737 F4WL95 A0A151WUX0 A0A151JF56 F4WL93 A0A1B0A3U4 B3MML8 E2BHE8 A0A026WE32 A0A310SQQ1 B4JC64
Pubmed
EMBL
KQ459078
KPJ03813.1
AGBW02010341
OWR48685.1
NWSH01000718
PCG74634.1
+ More
KQ459875 KPJ19597.1 GFDF01009085 JAV04999.1 GFDF01009100 JAV04984.1 AJVK01010728 AJVK01010729 KQ977349 KYN03409.1 ADTU01004604 ADTU01004605 ADTU01004606 GGFM01002147 MBW22898.1 AXCN02000024 CVRI01000055 CRL01200.1 KQ983238 KYQ46378.1 AJWK01011577 AJWK01011578 GDAI01000915 JAI16688.1 AAAB01008987 EAA01628.5 KK852806 KDR16091.1 AXCM01009254 GL888207 EGI65063.1 KQ971372 KYB25372.1 ADMH02002125 ETN58688.1 KYB25373.1 NNAY01000487 OXU28050.1 KQ414688 KOC63687.1 GARF01000045 JAC88903.1 GAMC01012082 JAB94473.1 GDHF01014132 JAI38182.1 GBXI01004707 JAD09585.1 GAKP01004984 GAKP01004982 JAC53970.1 JRES01001211 KNC24559.1 ATLV01012595 ATLV01012596 ATLV01012597 KE524806 KFB36910.1 GEDC01014139 JAS23159.1 KA645057 AFP59686.1 CH478294 EAT33290.1 KQ971371 EFA09999.1 GAMC01020936 JAB85619.1 GAMC01020938 JAB85617.1 KY849646 GAKP01018588 ATY74526.1 JAC40364.1 MH172531 QAA95938.1 GDHF01033733 GDHF01027985 JAI18581.1 JAI24329.1 KYN03410.1 JRES01000409 KNC31602.1 GFXV01005297 MBW17102.1 KP343867 AKJ85499.1 GBXI01004445 JAD09847.1 GBXI01005315 JAD08977.1 GL448287 EFN84918.1 JXJN01007808 ABLF02035201 ABLF02035202 DS235767 EEB16720.1 KZ149908 PZC78206.1 KQ980746 KYN13643.1 NWSH01002444 PCG68290.1 KA646285 AFP60914.1 KQ461003 KPJ10017.1 MF139835 ATY74536.1 JR052559 AEY61724.1 EEB16715.1 GBYB01008687 JAG78454.1 GFDL01000449 JAV34596.1 KYN03411.1 CH963857 EDW76377.1 KZ288474 PBC25293.1 KQ435812 KOX72863.1 EGI65061.1 OUUW01000010 SPP86037.1 KQ435180 KZC14992.1 GL768421 EFZ11364.1 EGI65058.1 KQ982717 KYQ51719.1 KQ979021 KYN24358.1 EGI65056.1 CH902620 EDV31909.1 KPU73697.1 EFN84917.1 KK107293 EZA53289.1 KQ760176 OAD61646.1 CH916368 EDW03077.1
KQ459875 KPJ19597.1 GFDF01009085 JAV04999.1 GFDF01009100 JAV04984.1 AJVK01010728 AJVK01010729 KQ977349 KYN03409.1 ADTU01004604 ADTU01004605 ADTU01004606 GGFM01002147 MBW22898.1 AXCN02000024 CVRI01000055 CRL01200.1 KQ983238 KYQ46378.1 AJWK01011577 AJWK01011578 GDAI01000915 JAI16688.1 AAAB01008987 EAA01628.5 KK852806 KDR16091.1 AXCM01009254 GL888207 EGI65063.1 KQ971372 KYB25372.1 ADMH02002125 ETN58688.1 KYB25373.1 NNAY01000487 OXU28050.1 KQ414688 KOC63687.1 GARF01000045 JAC88903.1 GAMC01012082 JAB94473.1 GDHF01014132 JAI38182.1 GBXI01004707 JAD09585.1 GAKP01004984 GAKP01004982 JAC53970.1 JRES01001211 KNC24559.1 ATLV01012595 ATLV01012596 ATLV01012597 KE524806 KFB36910.1 GEDC01014139 JAS23159.1 KA645057 AFP59686.1 CH478294 EAT33290.1 KQ971371 EFA09999.1 GAMC01020936 JAB85619.1 GAMC01020938 JAB85617.1 KY849646 GAKP01018588 ATY74526.1 JAC40364.1 MH172531 QAA95938.1 GDHF01033733 GDHF01027985 JAI18581.1 JAI24329.1 KYN03410.1 JRES01000409 KNC31602.1 GFXV01005297 MBW17102.1 KP343867 AKJ85499.1 GBXI01004445 JAD09847.1 GBXI01005315 JAD08977.1 GL448287 EFN84918.1 JXJN01007808 ABLF02035201 ABLF02035202 DS235767 EEB16720.1 KZ149908 PZC78206.1 KQ980746 KYN13643.1 NWSH01002444 PCG68290.1 KA646285 AFP60914.1 KQ461003 KPJ10017.1 MF139835 ATY74536.1 JR052559 AEY61724.1 EEB16715.1 GBYB01008687 JAG78454.1 GFDL01000449 JAV34596.1 KYN03411.1 CH963857 EDW76377.1 KZ288474 PBC25293.1 KQ435812 KOX72863.1 EGI65061.1 OUUW01000010 SPP86037.1 KQ435180 KZC14992.1 GL768421 EFZ11364.1 EGI65058.1 KQ982717 KYQ51719.1 KQ979021 KYN24358.1 EGI65056.1 CH902620 EDV31909.1 KPU73697.1 EFN84917.1 KK107293 EZA53289.1 KQ760176 OAD61646.1 CH916368 EDW03077.1
Proteomes
UP000053268
UP000007151
UP000218220
UP000053240
UP000092462
UP000078542
+ More
UP000075881 UP000075885 UP000076407 UP000005205 UP000075902 UP000075900 UP000076408 UP000075886 UP000075884 UP000183832 UP000075809 UP000092461 UP000075880 UP000007062 UP000027135 UP000069272 UP000075883 UP000007755 UP000075920 UP000002358 UP000192223 UP000007266 UP000000673 UP000215335 UP000053825 UP000095300 UP000037069 UP000030765 UP000095301 UP000008820 UP000091820 UP000092443 UP000008237 UP000092460 UP000007819 UP000092445 UP000009046 UP000078492 UP000078200 UP000007798 UP000242457 UP000053105 UP000268350 UP000076502 UP000007801 UP000053097 UP000001070
UP000075881 UP000075885 UP000076407 UP000005205 UP000075902 UP000075900 UP000076408 UP000075886 UP000075884 UP000183832 UP000075809 UP000092461 UP000075880 UP000007062 UP000027135 UP000069272 UP000075883 UP000007755 UP000075920 UP000002358 UP000192223 UP000007266 UP000000673 UP000215335 UP000053825 UP000095300 UP000037069 UP000030765 UP000095301 UP000008820 UP000091820 UP000092443 UP000008237 UP000092460 UP000007819 UP000092445 UP000009046 UP000078492 UP000078200 UP000007798 UP000242457 UP000053105 UP000268350 UP000076502 UP000007801 UP000053097 UP000001070
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0N1PG95
A0A212F4L4
A0A2A4JSP6
A0A194RQ20
A0A1L8DEW7
A0A1L8DF67
+ More
A0A1B0D2N6 A0A195CTG2 A0A182JPM5 A0A182P559 A0A182X0C0 A0A158NZH0 A0A182U3I1 A0A182R677 A0A182YAQ4 A0A2M3Z376 A0A182QE61 A0A182N442 A0A1J1IM12 A0A151WEU3 A0A1B0CGT5 A0A0K8TRC9 A0A182J8X7 Q7PYQ4 A0A067R2A0 A0A182FSU0 A0A182LV52 F4WLA0 A0A182VZ62 K7INT0 A0A1W4WF93 A0A139WBS7 W5J5L5 A0A139WBU4 A0A232FC36 A0A0L7QYQ1 K7J6H5 A0A1I8QDG6 A0A0U9HSE9 W8BBX8 A0A0K8VH42 A0A0A1XF64 A0A034WIK2 A0A0L0BZA0 A0A084VG16 A0A1B6DBU3 T1P8Q9 Q16GC7 D6X0P2 W8B731 W8AIT3 A0A034VCZ0 A0A3R5VSJ0 A0A0K8UCG2 A0A195CRZ4 A0A0L0CHB5 A0A1A9X055 A0A2H8TU48 A0A0G3FE97 A0A0A1XFV5 A0A1A9YT18 A0A0A1XDD6 E2BHE9 A0A1B0B3C6 J9K0F0 A0A1A9ZZ65 E0VTJ3 A0A2W1BZQ6 A0A151IYL4 A0A2A4JA19 A0A1I8PDS1 T1PDS1 A0A0N1INK1 A0A2H4TEV3 A0A1A9VFR8 V9IML8 E0VTI8 A0A0C9R7B6 A0A1Q3G4A7 A0A195CRT1 B4MV32 A0A2A3E0L0 A0A0M8ZZM1 F4WL98 A0A3B0JVB2 A0A154PSW7 E9J737 F4WL95 A0A151WUX0 A0A151JF56 F4WL93 A0A1B0A3U4 B3MML8 E2BHE8 A0A026WE32 A0A310SQQ1 B4JC64
A0A1B0D2N6 A0A195CTG2 A0A182JPM5 A0A182P559 A0A182X0C0 A0A158NZH0 A0A182U3I1 A0A182R677 A0A182YAQ4 A0A2M3Z376 A0A182QE61 A0A182N442 A0A1J1IM12 A0A151WEU3 A0A1B0CGT5 A0A0K8TRC9 A0A182J8X7 Q7PYQ4 A0A067R2A0 A0A182FSU0 A0A182LV52 F4WLA0 A0A182VZ62 K7INT0 A0A1W4WF93 A0A139WBS7 W5J5L5 A0A139WBU4 A0A232FC36 A0A0L7QYQ1 K7J6H5 A0A1I8QDG6 A0A0U9HSE9 W8BBX8 A0A0K8VH42 A0A0A1XF64 A0A034WIK2 A0A0L0BZA0 A0A084VG16 A0A1B6DBU3 T1P8Q9 Q16GC7 D6X0P2 W8B731 W8AIT3 A0A034VCZ0 A0A3R5VSJ0 A0A0K8UCG2 A0A195CRZ4 A0A0L0CHB5 A0A1A9X055 A0A2H8TU48 A0A0G3FE97 A0A0A1XFV5 A0A1A9YT18 A0A0A1XDD6 E2BHE9 A0A1B0B3C6 J9K0F0 A0A1A9ZZ65 E0VTJ3 A0A2W1BZQ6 A0A151IYL4 A0A2A4JA19 A0A1I8PDS1 T1PDS1 A0A0N1INK1 A0A2H4TEV3 A0A1A9VFR8 V9IML8 E0VTI8 A0A0C9R7B6 A0A1Q3G4A7 A0A195CRT1 B4MV32 A0A2A3E0L0 A0A0M8ZZM1 F4WL98 A0A3B0JVB2 A0A154PSW7 E9J737 F4WL95 A0A151WUX0 A0A151JF56 F4WL93 A0A1B0A3U4 B3MML8 E2BHE8 A0A026WE32 A0A310SQQ1 B4JC64
PDB
4YER
E-value=7.17849e-31,
Score=337
Ontologies
GO
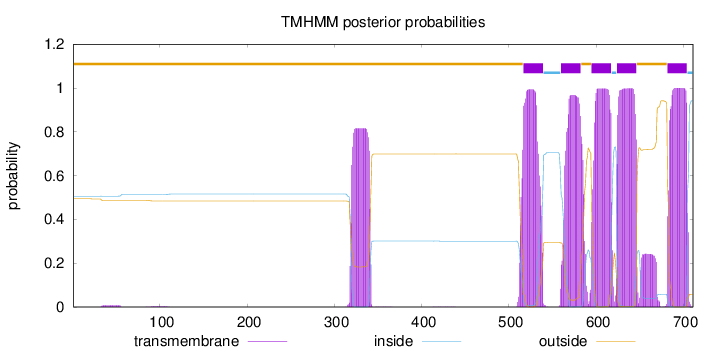
Topology
Length:
711
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
133.34662
Exp number, first 60 AAs:
0.17498
Total prob of N-in:
0.50619
outside
1 - 516
TMhelix
517 - 539
inside
540 - 559
TMhelix
560 - 582
outside
583 - 594
TMhelix
595 - 617
inside
618 - 623
TMhelix
624 - 646
outside
647 - 681
TMhelix
682 - 704
inside
705 - 711
Population Genetic Test Statistics
Pi
188.507165
Theta
153.155434
Tajima's D
0.317218
CLR
0.361732
CSRT
0.463276836158192
Interpretation
Uncertain
