Gene
KWMTBOMO15581
Pre Gene Modal
BGIBMGA010726
Annotation
PREDICTED:_ABC_transporter_G_family_member_20_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.634
Sequence
CDS
ATGCCAGCTTACGACAATGAAGGAGCCACGATATCCATGGAGGACGTCCGGGAAATCGACCGTCCAATCACCATTAGCACCATAGAAGAAGAGCTAATAGCCAGAAGAAACAGAATGTTCAAAACTCAAAAGTCCACAGTTGCTTCTAGGAGGCAGCAAGCGGTGTGTGTCAGACGAGCGCACAAAAGATATGGAACAGCTAAAAACCCTAACGTTATTTTGGATGGTCTGAATATGACTGTTGCTAAAGGATCTATATATGGCCTTTTGGGTGCTTCTGGCTGCGGCAAGACCACACTGCTGTCGTGTATAGTCGGCAGACGAAGACTAAATTCTGGTGAGATCTGGGTGCTGGGTGGAAAGCCCGGAAGCCCTGGCAGTGGAGTCCCTGGGCCTAGAATAGGATATATGCCACAGGAAGTCGCGCTCTTCGGCGAATTTTCGATCAGAGAAACACTAATATATTTCGGTTGGATCGCGAACATGTCCACTCCACAAGTAGAAGAACGAATAGACTTCCTCATCAAGTTATTGCAGTTACCTAATCCTACAAGACAAGTCAAAAATTTATCTGGGGGACAACAAAGAAGGACTTCTTTAGCCGCTGCCCTTATTCACGATCCTGAGCTGCTTATCCTGGACGAGCCTACAGTTGGAGTGGATCCTGTGCTCAGACAAAGCATCTGGGATCATCTCGTGGATATAACAAAGGAAGGTCGGACAACAGTGATCATAACTACTCATTACATCGACGAAACAAAACAAGCTAACATGATTGGCCTTATGCGAGGCGGTAGATTTTTGGCTGAAGAGTCTCCGAGCGAGTTGATCAGCCGCTACAACGCCGAAAGCCTTGAAGAAGTGTTCCTGAAGCTGTCAGTGCTACAGAATCTAGGCAAGCGCCGAAGATCTAGCATTCTCGCTGATGTCGTTGAAAAAGTTGAGCTTCCTACTATGCCTGATCCATCAGCCATAGATTTGGAGGAAGCAGAAGTAGGAGAGATTTCCGGAGAGTTCGGTGACAACGTCTCGATGAGCAGTAAAGGAAGGGTTGTAGTGGCACCGGACCTACCAATTCCAGTAGAAGCTATACCCCCTGAAGAGAAACCGAAGAGGACAAAAATGGAAATGATCATGCCAATGAAATGGCATCACATGAAAGCTCTAATATGGAAGAACTTCTGGAGTCTATTTAGGAACTTTGGTGTATTAGCGTTCATAATGGGCTTACCGATATCGCAAATGGTCTTATTCTGCCTCGCTATCGGTCATTATCCTGTTGGTCTTCCCATAGCGGTTGTGAATTACGAGTCTCACAGCAGCGATCCGTTGTGTTATTATGACCAGAAGAAATGCCCTCAAGACCGCAGCACGTACGATTGGCATTTGACGAACTTTAGCTGCCGTTATTTGGATTTCCTATCGAAGCGGCAGAGCAATCTGATCCTGTACCCCAACACTGAAGAAGCCATGTCGGAAGCAGCCCACGGCAAAGCCAGGGTCGTTCTGGTGTTCCCTGCCAATTTCTCAACTTGTCTTCAAGAAAGAATCACGAATCCACGCCGCACAGATGACTACACTATTATGAATTCCGATGTAGAGATCCACATGGATAATACAGATAAACAAGTCGAATGGATGTTGAGTCGCGATCTGCAGCTTGCGTTCATGGACGCTACACAGGAGCTGGCGAAGATCTGTCAGCTACCCTCGAGAATGATGTCCATTCCGGTGCACTTCAATGAGCCTATCTATGGTCTAGATGAGGCCAACTTCACCGATTTTGCTGGACCGGGAGTAATATTAACTATCATATTTTTCTTGGCGGTGGCTTTGACTTCAGGGTCTATGCTGGCTGAAAGAAATGAAGGCATCCTTGAAAGGACTCTAGTTTCAGGAATCACTGGTACAGAGATTCTGTTCTCACACGTCACGGTACAAATGCTGGTGATGATAGCCCAGTCGGTCATTGTCTTGGTCATCGGCTTCGTGTTCTTTGGACTCACGATGAGAGGTCCTATTGGATGGGTCACATTCCTCACACTGATCACTGGATTGTGTGGAATGACTTTTGGTTTCGTCGTGTCCTGTGTTTGTGACACCGACAGAACAGCTACTTACCTGGCTCTTGGCTCGTTCCTGCCGATGGTGTTACTTTGCGGCATCATATGGCCCGTTGAAGGTATGCACCTGATTCTACAGTGGATCAGCTACATTCTTCCGCTTACGAAATCTACAGAGTCTCTCAGATCGATGCTACAGAGAGGTTGGACTATCAATGTACCCACTGTCTACTCTGGATTCATATCAACCGGAATTTGGATAGCTATTTATTTGACCACAAGTATTTTACTACTTAAATTTAAGAAAGGCTAA
Protein
MPAYDNEGATISMEDVREIDRPITISTIEEELIARRNRMFKTQKSTVASRRQQAVCVRRAHKRYGTAKNPNVILDGLNMTVAKGSIYGLLGASGCGKTTLLSCIVGRRRLNSGEIWVLGGKPGSPGSGVPGPRIGYMPQEVALFGEFSIRETLIYFGWIANMSTPQVEERIDFLIKLLQLPNPTRQVKNLSGGQQRRTSLAAALIHDPELLILDEPTVGVDPVLRQSIWDHLVDITKEGRTTVIITTHYIDETKQANMIGLMRGGRFLAEESPSELISRYNAESLEEVFLKLSVLQNLGKRRRSSILADVVEKVELPTMPDPSAIDLEEAEVGEISGEFGDNVSMSSKGRVVVAPDLPIPVEAIPPEEKPKRTKMEMIMPMKWHHMKALIWKNFWSLFRNFGVLAFIMGLPISQMVLFCLAIGHYPVGLPIAVVNYESHSSDPLCYYDQKKCPQDRSTYDWHLTNFSCRYLDFLSKRQSNLILYPNTEEAMSEAAHGKARVVLVFPANFSTCLQERITNPRRTDDYTIMNSDVEIHMDNTDKQVEWMLSRDLQLAFMDATQELAKICQLPSRMMSIPVHFNEPIYGLDEANFTDFAGPGVILTIIFFLAVALTSGSMLAERNEGILERTLVSGITGTEILFSHVTVQMLVMIAQSVIVLVIGFVFFGLTMRGPIGWVTFLTLITGLCGMTFGFVVSCVCDTDRTATYLALGSFLPMVLLCGIIWPVEGMHLILQWISYILPLTKSTESLRSMLQRGWTINVPTVYSGFISTGIWIAIYLTTSILLLKFKKG
Summary
Uniprot
H9JMH3
A0A2H1V7V2
A0A0N1IMY5
A0A0N1IQR3
A0A212F4U8
A0A182JL24
+ More
W5JPR1 A0A084WUS8 A0A182FPA4 A0A2M4BEJ1 A0A182P3S9 A0A2M4AFI7 A0A182QMX6 A0A182Y6M8 A0A182M5B0 A0A182LEG1 Q7QCI0 A0A182HSF4 A0A182UF67 A0A182V3M4 A0A182XAL3 A0A182RQH9 A0A182W0R8 A0A182K5K8 A0A1S4FAP5 Q179W7 A0A023EW46 A0A182NRC8 A0A1J1IC19 A0A1Q3FPY4 A0A1I8PN83 D6X2Z9 T1PHQ0 A0A0L0CB18 A0A0K8UA84 B3P683 B4PSU8 A0A0A1X0R1 B5DVI8 B4GED2 Q86P18 W8AXS4 B3N1L9 B4NKQ9 A0A3B0JM23 A0A0M4F3C0 B4QW42 B4LYD6 A0A2H8TVP8 A0A3R5SLG5 A0A1L8DDG1 E2BHE8 A0A151JF56 B4KAL2 A0A151WUX0 A0A1W4VLH2 A0A154PSW7 B4ICL0 A0A310SQQ1 A0A195FCD0 A0A0L7QLD0 F4WL93 A0A336MFA5 A0A1B6LIH2 A0A195BMU2 A0A026WE32 A0A1B6DVQ2 A0A151I6F8 J9K0T3 A0A232FG17 A0A0J7KE16 N6UEZ5 J3JUX4 V9IML8 A0A1B6GX14 A0A1B0GF05 A0A0K8TMQ7 A0A1B0GF04 A0A2A3E0L0 A0A0C9R254 A0A0M8ZZM1 B0WZM5 A0A0K8S4N7 A0A146LDF7 A0A161ANH5 A0A1Y1KEG6 A0A0A9Z6F2 E2AB08 A0A2J7PYS4 A0A069DWS8 A0A023F3A5 A0A0V0G456 A0A3L8DRF1 A0A0L7LKY8 A0A224X869 A0A0A9YYT0 A0A067QZX9 A0A182H1D3 A0A139WBU4
W5JPR1 A0A084WUS8 A0A182FPA4 A0A2M4BEJ1 A0A182P3S9 A0A2M4AFI7 A0A182QMX6 A0A182Y6M8 A0A182M5B0 A0A182LEG1 Q7QCI0 A0A182HSF4 A0A182UF67 A0A182V3M4 A0A182XAL3 A0A182RQH9 A0A182W0R8 A0A182K5K8 A0A1S4FAP5 Q179W7 A0A023EW46 A0A182NRC8 A0A1J1IC19 A0A1Q3FPY4 A0A1I8PN83 D6X2Z9 T1PHQ0 A0A0L0CB18 A0A0K8UA84 B3P683 B4PSU8 A0A0A1X0R1 B5DVI8 B4GED2 Q86P18 W8AXS4 B3N1L9 B4NKQ9 A0A3B0JM23 A0A0M4F3C0 B4QW42 B4LYD6 A0A2H8TVP8 A0A3R5SLG5 A0A1L8DDG1 E2BHE8 A0A151JF56 B4KAL2 A0A151WUX0 A0A1W4VLH2 A0A154PSW7 B4ICL0 A0A310SQQ1 A0A195FCD0 A0A0L7QLD0 F4WL93 A0A336MFA5 A0A1B6LIH2 A0A195BMU2 A0A026WE32 A0A1B6DVQ2 A0A151I6F8 J9K0T3 A0A232FG17 A0A0J7KE16 N6UEZ5 J3JUX4 V9IML8 A0A1B6GX14 A0A1B0GF05 A0A0K8TMQ7 A0A1B0GF04 A0A2A3E0L0 A0A0C9R254 A0A0M8ZZM1 B0WZM5 A0A0K8S4N7 A0A146LDF7 A0A161ANH5 A0A1Y1KEG6 A0A0A9Z6F2 E2AB08 A0A2J7PYS4 A0A069DWS8 A0A023F3A5 A0A0V0G456 A0A3L8DRF1 A0A0L7LKY8 A0A224X869 A0A0A9YYT0 A0A067QZX9 A0A182H1D3 A0A139WBU4
Pubmed
19121390
26354079
22118469
20920257
23761445
24438588
+ More
25244985 20966253 12364791 14747013 17210077 17510324 24945155 18362917 19820115 25315136 26108605 17994087 17550304 25830018 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 20798317 21719571 24508170 28648823 23537049 22516182 26369729 26823975 28004739 25401762 26334808 25474469 30249741 26227816 24845553 26483478
25244985 20966253 12364791 14747013 17210077 17510324 24945155 18362917 19820115 25315136 26108605 17994087 17550304 25830018 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 20798317 21719571 24508170 28648823 23537049 22516182 26369729 26823975 28004739 25401762 26334808 25474469 30249741 26227816 24845553 26483478
EMBL
BABH01016791
BABH01016792
ODYU01001121
SOQ36891.1
KQ459078
KPJ03812.1
+ More
LADJ01026996 KPJ21158.1 AGBW02010306 OWR48761.1 ADMH02000693 ETN65298.1 ATLV01027119 KE525423 KFB53972.1 GGFJ01002329 MBW51470.1 GGFK01006087 MBW39408.1 AXCN02001466 AXCM01000158 AAAB01008859 EAA08206.4 APCN01000271 CH477344 EAT43031.1 GAPW01000262 JAC13336.1 CVRI01000047 CRK97747.1 GFDL01005512 JAV29533.1 KQ971372 EFA10297.1 KA647650 AFP62279.1 JRES01000664 KNC29441.1 GDHF01028806 JAI23508.1 CH954182 EDV53553.1 CM000160 EDW98635.1 GBXI01010069 GBXI01008924 GBXI01007441 GBXI01000817 JAD04223.1 JAD05368.1 JAD06851.1 JAD13475.1 CM000070 EDY68204.2 CH479182 EDW33967.1 BT003531 AE014297 AAO39535.1 ABC66191.1 GAMC01012770 GAMC01012769 JAB93785.1 CH902661 EDV33988.1 CH964272 EDW84120.1 OUUW01000007 SPP83285.1 CP012526 ALC45695.1 CM000364 EDX14541.1 CH940650 EDW66932.1 GFXV01006502 MBW18307.1 MH172535 QAA95942.1 GFDF01009575 JAV04509.1 GL448287 EFN84917.1 KQ979021 KYN24358.1 CH933806 EDW15725.2 KQ982717 KYQ51719.1 KQ435180 KZC14992.1 CH480828 EDW45106.1 KQ760176 OAD61646.1 KQ981685 KYN38051.1 KQ414915 KOC59442.1 GL888207 EGI65056.1 UFQT01000937 SSX28081.1 GEBQ01016518 JAT23459.1 KQ976433 KYM87308.1 KK107293 EZA53289.1 GEDC01007530 JAS29768.1 KQ978473 KYM93686.1 ABLF02033861 NNAY01000251 OXU29694.1 LBMM01009052 KMQ88436.1 APGK01029562 KB740735 KB632302 ENN79196.1 ERL91767.1 BT127039 AEE62001.1 JR052559 AEY61724.1 GECZ01002834 JAS66935.1 CCAG010012701 CCAG010012702 CCAG010012703 GDAI01001971 JAI15632.1 KZ288474 PBC25293.1 GBYB01000940 GBYB01000941 JAG70707.1 JAG70708.1 KQ435812 KOX72863.1 DS232211 EDS37539.1 GBRD01017619 JAG48208.1 GDHC01012408 JAQ06221.1 KF828807 AIN44124.1 GEZM01086010 JAV59839.1 GBHO01006224 JAG37380.1 GL438234 EFN69284.1 NEVH01020342 PNF21462.1 GBGD01000603 JAC88286.1 GBBI01002817 JAC15895.1 GECL01003381 JAP02743.1 QOIP01000005 RLU22987.1 JTDY01000691 KOB76203.1 GFTR01007880 JAW08546.1 GBHO01006230 JAG37374.1 KK852806 KDR16094.1 JXUM01003433 KQ560158 KXJ84121.1 KYB25373.1
LADJ01026996 KPJ21158.1 AGBW02010306 OWR48761.1 ADMH02000693 ETN65298.1 ATLV01027119 KE525423 KFB53972.1 GGFJ01002329 MBW51470.1 GGFK01006087 MBW39408.1 AXCN02001466 AXCM01000158 AAAB01008859 EAA08206.4 APCN01000271 CH477344 EAT43031.1 GAPW01000262 JAC13336.1 CVRI01000047 CRK97747.1 GFDL01005512 JAV29533.1 KQ971372 EFA10297.1 KA647650 AFP62279.1 JRES01000664 KNC29441.1 GDHF01028806 JAI23508.1 CH954182 EDV53553.1 CM000160 EDW98635.1 GBXI01010069 GBXI01008924 GBXI01007441 GBXI01000817 JAD04223.1 JAD05368.1 JAD06851.1 JAD13475.1 CM000070 EDY68204.2 CH479182 EDW33967.1 BT003531 AE014297 AAO39535.1 ABC66191.1 GAMC01012770 GAMC01012769 JAB93785.1 CH902661 EDV33988.1 CH964272 EDW84120.1 OUUW01000007 SPP83285.1 CP012526 ALC45695.1 CM000364 EDX14541.1 CH940650 EDW66932.1 GFXV01006502 MBW18307.1 MH172535 QAA95942.1 GFDF01009575 JAV04509.1 GL448287 EFN84917.1 KQ979021 KYN24358.1 CH933806 EDW15725.2 KQ982717 KYQ51719.1 KQ435180 KZC14992.1 CH480828 EDW45106.1 KQ760176 OAD61646.1 KQ981685 KYN38051.1 KQ414915 KOC59442.1 GL888207 EGI65056.1 UFQT01000937 SSX28081.1 GEBQ01016518 JAT23459.1 KQ976433 KYM87308.1 KK107293 EZA53289.1 GEDC01007530 JAS29768.1 KQ978473 KYM93686.1 ABLF02033861 NNAY01000251 OXU29694.1 LBMM01009052 KMQ88436.1 APGK01029562 KB740735 KB632302 ENN79196.1 ERL91767.1 BT127039 AEE62001.1 JR052559 AEY61724.1 GECZ01002834 JAS66935.1 CCAG010012701 CCAG010012702 CCAG010012703 GDAI01001971 JAI15632.1 KZ288474 PBC25293.1 GBYB01000940 GBYB01000941 JAG70707.1 JAG70708.1 KQ435812 KOX72863.1 DS232211 EDS37539.1 GBRD01017619 JAG48208.1 GDHC01012408 JAQ06221.1 KF828807 AIN44124.1 GEZM01086010 JAV59839.1 GBHO01006224 JAG37380.1 GL438234 EFN69284.1 NEVH01020342 PNF21462.1 GBGD01000603 JAC88286.1 GBBI01002817 JAC15895.1 GECL01003381 JAP02743.1 QOIP01000005 RLU22987.1 JTDY01000691 KOB76203.1 GFTR01007880 JAW08546.1 GBHO01006230 JAG37374.1 KK852806 KDR16094.1 JXUM01003433 KQ560158 KXJ84121.1 KYB25373.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000075880
UP000000673
+ More
UP000030765 UP000069272 UP000075885 UP000075886 UP000076408 UP000075883 UP000075882 UP000007062 UP000075840 UP000075902 UP000075903 UP000076407 UP000075900 UP000075920 UP000075881 UP000008820 UP000075884 UP000183832 UP000095300 UP000007266 UP000095301 UP000037069 UP000008711 UP000002282 UP000001819 UP000008744 UP000000803 UP000007801 UP000007798 UP000268350 UP000092553 UP000000304 UP000008792 UP000008237 UP000078492 UP000009192 UP000075809 UP000192221 UP000076502 UP000001292 UP000078541 UP000053825 UP000007755 UP000078540 UP000053097 UP000078542 UP000007819 UP000215335 UP000036403 UP000019118 UP000030742 UP000092444 UP000242457 UP000053105 UP000002320 UP000000311 UP000235965 UP000279307 UP000037510 UP000027135 UP000069940 UP000249989
UP000030765 UP000069272 UP000075885 UP000075886 UP000076408 UP000075883 UP000075882 UP000007062 UP000075840 UP000075902 UP000075903 UP000076407 UP000075900 UP000075920 UP000075881 UP000008820 UP000075884 UP000183832 UP000095300 UP000007266 UP000095301 UP000037069 UP000008711 UP000002282 UP000001819 UP000008744 UP000000803 UP000007801 UP000007798 UP000268350 UP000092553 UP000000304 UP000008792 UP000008237 UP000078492 UP000009192 UP000075809 UP000192221 UP000076502 UP000001292 UP000078541 UP000053825 UP000007755 UP000078540 UP000053097 UP000078542 UP000007819 UP000215335 UP000036403 UP000019118 UP000030742 UP000092444 UP000242457 UP000053105 UP000002320 UP000000311 UP000235965 UP000279307 UP000037510 UP000027135 UP000069940 UP000249989
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9JMH3
A0A2H1V7V2
A0A0N1IMY5
A0A0N1IQR3
A0A212F4U8
A0A182JL24
+ More
W5JPR1 A0A084WUS8 A0A182FPA4 A0A2M4BEJ1 A0A182P3S9 A0A2M4AFI7 A0A182QMX6 A0A182Y6M8 A0A182M5B0 A0A182LEG1 Q7QCI0 A0A182HSF4 A0A182UF67 A0A182V3M4 A0A182XAL3 A0A182RQH9 A0A182W0R8 A0A182K5K8 A0A1S4FAP5 Q179W7 A0A023EW46 A0A182NRC8 A0A1J1IC19 A0A1Q3FPY4 A0A1I8PN83 D6X2Z9 T1PHQ0 A0A0L0CB18 A0A0K8UA84 B3P683 B4PSU8 A0A0A1X0R1 B5DVI8 B4GED2 Q86P18 W8AXS4 B3N1L9 B4NKQ9 A0A3B0JM23 A0A0M4F3C0 B4QW42 B4LYD6 A0A2H8TVP8 A0A3R5SLG5 A0A1L8DDG1 E2BHE8 A0A151JF56 B4KAL2 A0A151WUX0 A0A1W4VLH2 A0A154PSW7 B4ICL0 A0A310SQQ1 A0A195FCD0 A0A0L7QLD0 F4WL93 A0A336MFA5 A0A1B6LIH2 A0A195BMU2 A0A026WE32 A0A1B6DVQ2 A0A151I6F8 J9K0T3 A0A232FG17 A0A0J7KE16 N6UEZ5 J3JUX4 V9IML8 A0A1B6GX14 A0A1B0GF05 A0A0K8TMQ7 A0A1B0GF04 A0A2A3E0L0 A0A0C9R254 A0A0M8ZZM1 B0WZM5 A0A0K8S4N7 A0A146LDF7 A0A161ANH5 A0A1Y1KEG6 A0A0A9Z6F2 E2AB08 A0A2J7PYS4 A0A069DWS8 A0A023F3A5 A0A0V0G456 A0A3L8DRF1 A0A0L7LKY8 A0A224X869 A0A0A9YYT0 A0A067QZX9 A0A182H1D3 A0A139WBU4
W5JPR1 A0A084WUS8 A0A182FPA4 A0A2M4BEJ1 A0A182P3S9 A0A2M4AFI7 A0A182QMX6 A0A182Y6M8 A0A182M5B0 A0A182LEG1 Q7QCI0 A0A182HSF4 A0A182UF67 A0A182V3M4 A0A182XAL3 A0A182RQH9 A0A182W0R8 A0A182K5K8 A0A1S4FAP5 Q179W7 A0A023EW46 A0A182NRC8 A0A1J1IC19 A0A1Q3FPY4 A0A1I8PN83 D6X2Z9 T1PHQ0 A0A0L0CB18 A0A0K8UA84 B3P683 B4PSU8 A0A0A1X0R1 B5DVI8 B4GED2 Q86P18 W8AXS4 B3N1L9 B4NKQ9 A0A3B0JM23 A0A0M4F3C0 B4QW42 B4LYD6 A0A2H8TVP8 A0A3R5SLG5 A0A1L8DDG1 E2BHE8 A0A151JF56 B4KAL2 A0A151WUX0 A0A1W4VLH2 A0A154PSW7 B4ICL0 A0A310SQQ1 A0A195FCD0 A0A0L7QLD0 F4WL93 A0A336MFA5 A0A1B6LIH2 A0A195BMU2 A0A026WE32 A0A1B6DVQ2 A0A151I6F8 J9K0T3 A0A232FG17 A0A0J7KE16 N6UEZ5 J3JUX4 V9IML8 A0A1B6GX14 A0A1B0GF05 A0A0K8TMQ7 A0A1B0GF04 A0A2A3E0L0 A0A0C9R254 A0A0M8ZZM1 B0WZM5 A0A0K8S4N7 A0A146LDF7 A0A161ANH5 A0A1Y1KEG6 A0A0A9Z6F2 E2AB08 A0A2J7PYS4 A0A069DWS8 A0A023F3A5 A0A0V0G456 A0A3L8DRF1 A0A0L7LKY8 A0A224X869 A0A0A9YYT0 A0A067QZX9 A0A182H1D3 A0A139WBU4
PDB
4YER
E-value=1.27885e-25,
Score=292
Ontologies
GO
Topology
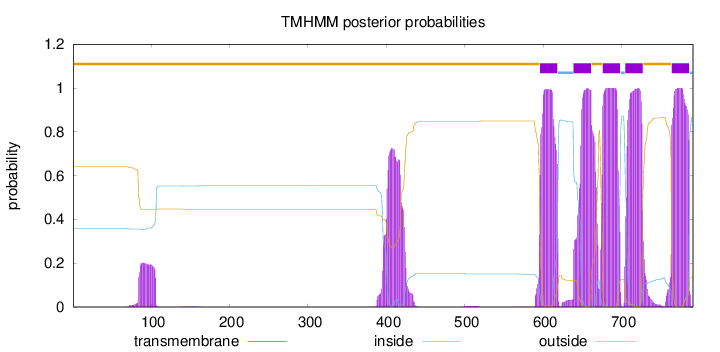
Length:
791
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
134.83892
Exp number, first 60 AAs:
0
Total prob of N-in:
0.35814
outside
1 - 595
TMhelix
596 - 618
inside
619 - 638
TMhelix
639 - 661
outside
662 - 675
TMhelix
676 - 698
inside
699 - 704
TMhelix
705 - 727
outside
728 - 763
TMhelix
764 - 786
inside
787 - 791
Population Genetic Test Statistics
Pi
256.966313
Theta
23.738279
Tajima's D
0.043394
CLR
0.738173
CSRT
0.390530473476326
Interpretation
Uncertain
