Gene
KWMTBOMO15580
Pre Gene Modal
BGIBMGA010823
Annotation
PREDICTED:_lysyl_oxidase_homolog_3_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 2.781
Sequence
CDS
ATGAAAAAGCATCTAGTGTTTAATTCCACATTGCTGTTTGTAATGTTGATCGAATCTATTGGAGTTTCGGGCAAAGATTTGAGTGAAAATGAGCGGGCTGGCCGAGCGGCATTTGTTCAAAGACTATTGAATAAAAAAAAGTATTTGGAAGGGAGAGTGAAATTAGTTGGCGGGAATAAATATGAAGGCAACGTGTACATCTATCACGCCGGTCGTTGGGGAGCTATTTGTGACGACAGCTGGGATGATGCGGCAGCTCAAATCGTATGTCGCGTGTTCAACAGGACTGGTTTTGCTACTCACGGAGGGCAATACGGTGAAGCAAAAGAACAATTTTGGATGGATGACATGGTTTGTCAGGGTGATGAATCTTCACTGACCCAGTGCATATTCTCTGGCTGGGGTTCTTCAGACTGCACGCCTACTGAAGCAGCTGGAGTCAAATGCCGCAAACAAGAGACACATGTGCCATTAAATCTTTCCTCAAAGAAAAAAGCAGGAGTCCCACTACATGAAGTCTTAGATATTAGGTATACTAGTTTGAGACTAGTTGGCGGAAGAAATGCTCTGGAGGGCAGAGTGGAGATCTATCACAATAATTACTGGGGAAGCCTATGCCCAGACGGCTGGACGATCTATGAAGCTGCAGCTACCTGTCGCCATTTAGGCCTAGGATATGCGGAGCAAGCCCTACAGACGGATTATTTCGGAAGCTCAAAGATTGTTCTTAGCGGAGTGGAATGTCAAGGCAACGAGACGAACCTATTAATGTGTCGACATCAGAAATATGGCAACGTTACGTGCCCAGGGACAGTTGGTCACGTCGCAGCGGTAATATGTACCCAAAAATTGGCCGATTTGGAGCTTGACACTGCAGCCATAGAACACACCGCCCATTTACAAGACGTTCCAATGTACCAGCTCACTTGCGCTATGGAAGAAAACTGCTTGACGAAGTCAGCCTATGAGACGCAAAAAACCAACCCAAATTGGCAATATGAAAGCAGAAGGCTATTGCGTTTCACAGCAGCCTCTCTCAACATCGGAAACGATGAATTCAGGCCGTATTTACCGAAGCATTTGTGGCAATGGCATCTTTGTCATATGCACTACCACAGTATGGAAGTGTTTGCTACTTTCGATGTCTTCGATGCGTCAGGCCGAAGAGTCGCAGAAGGCCATAAAGCTTCGTTCTGCTTAGAGGATAACACGTGCATGCCTGGAGTGGAAAAAAAGTATTCCTGCAAGAACTATGGAGATCAAGGTGTTTCAGTGAACTGTTCGGATGTTTATCATTACAATATTGACTGTCAGTGGGTGGATGTGAGTGATGTGGAGCCAGGCGATTACACGTTAAAGGTGGCAGTGAACCCTCACGCGCGAATAGCAGAACAAAATTACCACAATAACGCGGCGTCTTGCAGACTGCGTCTTTCTGAACTCTATGCTGCAGTATACGGATGTAAACGCGAAAGGCCTTAG
Protein
MKKHLVFNSTLLFVMLIESIGVSGKDLSENERAGRAAFVQRLLNKKKYLEGRVKLVGGNKYEGNVYIYHAGRWGAICDDSWDDAAAQIVCRVFNRTGFATHGGQYGEAKEQFWMDDMVCQGDESSLTQCIFSGWGSSDCTPTEAAGVKCRKQETHVPLNLSSKKKAGVPLHEVLDIRYTSLRLVGGRNALEGRVEIYHNNYWGSLCPDGWTIYEAAATCRHLGLGYAEQALQTDYFGSSKIVLSGVECQGNETNLLMCRHQKYGNVTCPGTVGHVAAVICTQKLADLELDTAAIEHTAHLQDVPMYQLTCAMEENCLTKSAYETQKTNPNWQYESRRLLRFTAASLNIGNDEFRPYLPKHLWQWHLCHMHYHSMEVFATFDVFDASGRRVAEGHKASFCLEDNTCMPGVEKKYSCKNYGDQGVSVNCSDVYHYNIDCQWVDVSDVEPGDYTLKVAVNPHARIAEQNYHNNAASCRLRLSELYAAVYGCKRERP
Summary
Uniprot
H9JMS0
A0A0N1PI89
A0A0N1I3I6
A0A3S2NN45
A0A2H1V7S0
A0A2A4J826
+ More
A0A212F4W5 A0A084W394 A0A182JJM3 A0A182N5M8 A0A182QRR0 A0A182YGH7 A0A182MNL6 A0A182W0I7 Q7QBA0 A0A182L7I8 A0A182I7X8 B4P8E9 B3MJ04 A0A182VAU7 A0A0L0CM11 A0A182RYI1 A0A182TK12 B3NN17 A0A182XIX9 A0A182F6N6 B4GHF6 A0A182P8X5 Q28XR1 T1PJI8 B4I7W2 A0A3L8DKP5 A0A1W4UZ01 A0A0J9U811 B4MS10 B4QGL0 A0A1I8MTI8 A0A195D5J7 E2BNK3 B0WJ73 E2AIQ3 A0A151WMF5 A0A3B0JP91 Q9W2C9 H0RNL9 A0A1Q3FPA5 A0A034WB96 A0A1A9Y7Y6 A0A1B0ARI1 F4WLF0 A0A1J1I5J6 A0A0M4E8C4 W5J9E1 Q9N9Y8 A0A2J7PI67 A0A1A9ZRG9 A0A026X3Y3 A0A1A9X3F4 A0A195FY20 B4J6A1 A0A034W761 A0A1I8PDV5 A0A158NVI9 A0A0A1WG43 A0A182H0M1 A0A154NVR5 B4LLQ2 W8C566 C5WLU2 V9IFH6 E9IWS9 B4KNS1 A0A1Y1MMJ8 D6X0V2 A0A0L7RJH8 A0A0J7L8M1 E0VBQ5 A0A0K8TZK6 A0A195BMZ9 A0A1A9VEE9 K7IM98 A0A087TCQ9 E9GHI3 A0A088ADF0 A0A195E561 A0A067R7D2 A0A0P5YL50 A0A0P5EIX5 A0A0N8AL55 A0A0P6AM69 T1E173 A0A0N0U4V5 A0A0N8E1K9 A0A147BK26 A0A0P5YQE2 A0A2H8TD94 N6TMA4 A0A0P5XQY5 A0A1W4XWS8 A0A2S2Q7B9
A0A212F4W5 A0A084W394 A0A182JJM3 A0A182N5M8 A0A182QRR0 A0A182YGH7 A0A182MNL6 A0A182W0I7 Q7QBA0 A0A182L7I8 A0A182I7X8 B4P8E9 B3MJ04 A0A182VAU7 A0A0L0CM11 A0A182RYI1 A0A182TK12 B3NN17 A0A182XIX9 A0A182F6N6 B4GHF6 A0A182P8X5 Q28XR1 T1PJI8 B4I7W2 A0A3L8DKP5 A0A1W4UZ01 A0A0J9U811 B4MS10 B4QGL0 A0A1I8MTI8 A0A195D5J7 E2BNK3 B0WJ73 E2AIQ3 A0A151WMF5 A0A3B0JP91 Q9W2C9 H0RNL9 A0A1Q3FPA5 A0A034WB96 A0A1A9Y7Y6 A0A1B0ARI1 F4WLF0 A0A1J1I5J6 A0A0M4E8C4 W5J9E1 Q9N9Y8 A0A2J7PI67 A0A1A9ZRG9 A0A026X3Y3 A0A1A9X3F4 A0A195FY20 B4J6A1 A0A034W761 A0A1I8PDV5 A0A158NVI9 A0A0A1WG43 A0A182H0M1 A0A154NVR5 B4LLQ2 W8C566 C5WLU2 V9IFH6 E9IWS9 B4KNS1 A0A1Y1MMJ8 D6X0V2 A0A0L7RJH8 A0A0J7L8M1 E0VBQ5 A0A0K8TZK6 A0A195BMZ9 A0A1A9VEE9 K7IM98 A0A087TCQ9 E9GHI3 A0A088ADF0 A0A195E561 A0A067R7D2 A0A0P5YL50 A0A0P5EIX5 A0A0N8AL55 A0A0P6AM69 T1E173 A0A0N0U4V5 A0A0N8E1K9 A0A147BK26 A0A0P5YQE2 A0A2H8TD94 N6TMA4 A0A0P5XQY5 A0A1W4XWS8 A0A2S2Q7B9
Pubmed
19121390
26354079
22118469
24438588
25244985
12364791
+ More
14747013 17210077 20966253 17994087 17550304 26108605 15632085 30249741 22936249 25315136 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25348373 21719571 20920257 23761445 12686136 15811848 24508170 21347285 25830018 26483478 24495485 21282665 28004739 18362917 19820115 20566863 20075255 21292972 24845553 29652888 23537049
14747013 17210077 20966253 17994087 17550304 26108605 15632085 30249741 22936249 25315136 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25348373 21719571 20920257 23761445 12686136 15811848 24508170 21347285 25830018 26483478 24495485 21282665 28004739 18362917 19820115 20566863 20075255 21292972 24845553 29652888 23537049
EMBL
BABH01016791
LADJ01026996
KPJ21157.1
KQ459078
KPJ03811.1
RSAL01000221
+ More
RVE44046.1 ODYU01001122 SOQ36895.1 NWSH01002740 PCG67600.1 AGBW02010306 OWR48762.1 ATLV01019867 ATLV01019868 ATLV01019869 KE525281 KFB44688.1 AXCN02001159 AXCM01001728 AAAB01008880 EAA08587.1 APCN01002579 CM000158 EDW91189.1 CH902619 EDV37070.1 JRES01000195 KNC33373.1 CH954179 EDV55511.1 CH479183 EDW35926.1 CM000071 EAL26255.2 KA648093 AFP62722.1 CH480824 EDW56687.1 QOIP01000007 RLU20408.1 CM002911 KMY95660.1 CH963850 EDW74899.2 CM000362 EDX08129.1 KQ976870 KYN07714.1 GL449414 EFN82782.1 DS231956 EDS28963.1 GL439853 EFN66690.1 KQ982944 KYQ49049.1 OUUW01000001 SPP75419.1 AE013599 AAF46763.1 BT132941 AEW26265.1 GFDL01005729 JAV29316.1 GAKP01007542 JAC51410.1 JXJN01002447 GL888207 EGI65113.1 CVRI01000037 CRK93633.1 CP012524 ALC40946.1 ADMH02001794 ETN61077.1 AJ295626 CAB99481.1 NEVH01025129 PNF16040.1 KK107019 EZA62803.1 KQ981169 KYN45338.1 CH916367 EDW00874.1 GAKP01007541 JAC51411.1 ADTU01027213 GBXI01016626 JAC97665.1 JXUM01101570 JXUM01101571 JXUM01101572 JXUM01101573 KQ564658 KXJ71944.1 KQ434766 KZC03737.1 CH940648 EDW60915.1 GAMC01005074 JAC01482.1 BT088907 ACT09404.1 JR040656 AEY59226.1 GL766597 EFZ15003.1 CH933808 EDW10056.1 GEZM01030958 JAV85136.1 KQ971372 EFA10680.1 KQ414581 KOC71030.1 LBMM01000238 KMR03324.1 DS235035 EEB10811.1 GDHF01032400 JAI19914.1 KQ976433 KYM87355.1 KK114620 KFM62898.1 GL732545 EFX81056.1 KQ979657 KYN19989.1 KK852895 KDR14196.1 GDIP01056252 LRGB01002580 JAM47463.1 KZS06844.1 GDIQ01268882 JAJ82842.1 GDIQ01265019 JAJ86705.1 GDIP01027357 JAM76358.1 GAKT01000144 JAA92918.1 KQ435812 KOX72728.1 GDIQ01070796 JAN23941.1 GEGO01004261 JAR91143.1 GDIP01054713 JAM49002.1 GFXV01000276 MBW12081.1 APGK01059051 KB741292 ENN70385.1 GDIP01080941 JAM22774.1 GGMS01003889 MBY73092.1
RVE44046.1 ODYU01001122 SOQ36895.1 NWSH01002740 PCG67600.1 AGBW02010306 OWR48762.1 ATLV01019867 ATLV01019868 ATLV01019869 KE525281 KFB44688.1 AXCN02001159 AXCM01001728 AAAB01008880 EAA08587.1 APCN01002579 CM000158 EDW91189.1 CH902619 EDV37070.1 JRES01000195 KNC33373.1 CH954179 EDV55511.1 CH479183 EDW35926.1 CM000071 EAL26255.2 KA648093 AFP62722.1 CH480824 EDW56687.1 QOIP01000007 RLU20408.1 CM002911 KMY95660.1 CH963850 EDW74899.2 CM000362 EDX08129.1 KQ976870 KYN07714.1 GL449414 EFN82782.1 DS231956 EDS28963.1 GL439853 EFN66690.1 KQ982944 KYQ49049.1 OUUW01000001 SPP75419.1 AE013599 AAF46763.1 BT132941 AEW26265.1 GFDL01005729 JAV29316.1 GAKP01007542 JAC51410.1 JXJN01002447 GL888207 EGI65113.1 CVRI01000037 CRK93633.1 CP012524 ALC40946.1 ADMH02001794 ETN61077.1 AJ295626 CAB99481.1 NEVH01025129 PNF16040.1 KK107019 EZA62803.1 KQ981169 KYN45338.1 CH916367 EDW00874.1 GAKP01007541 JAC51411.1 ADTU01027213 GBXI01016626 JAC97665.1 JXUM01101570 JXUM01101571 JXUM01101572 JXUM01101573 KQ564658 KXJ71944.1 KQ434766 KZC03737.1 CH940648 EDW60915.1 GAMC01005074 JAC01482.1 BT088907 ACT09404.1 JR040656 AEY59226.1 GL766597 EFZ15003.1 CH933808 EDW10056.1 GEZM01030958 JAV85136.1 KQ971372 EFA10680.1 KQ414581 KOC71030.1 LBMM01000238 KMR03324.1 DS235035 EEB10811.1 GDHF01032400 JAI19914.1 KQ976433 KYM87355.1 KK114620 KFM62898.1 GL732545 EFX81056.1 KQ979657 KYN19989.1 KK852895 KDR14196.1 GDIP01056252 LRGB01002580 JAM47463.1 KZS06844.1 GDIQ01268882 JAJ82842.1 GDIQ01265019 JAJ86705.1 GDIP01027357 JAM76358.1 GAKT01000144 JAA92918.1 KQ435812 KOX72728.1 GDIQ01070796 JAN23941.1 GEGO01004261 JAR91143.1 GDIP01054713 JAM49002.1 GFXV01000276 MBW12081.1 APGK01059051 KB741292 ENN70385.1 GDIP01080941 JAM22774.1 GGMS01003889 MBY73092.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000030765 UP000075880 UP000075884 UP000075886 UP000076408 UP000075883 UP000075920 UP000007062 UP000075882 UP000075840 UP000002282 UP000007801 UP000075903 UP000037069 UP000075900 UP000075902 UP000008711 UP000076407 UP000069272 UP000008744 UP000075885 UP000001819 UP000001292 UP000279307 UP000192221 UP000007798 UP000000304 UP000095301 UP000078542 UP000008237 UP000002320 UP000000311 UP000075809 UP000268350 UP000000803 UP000092443 UP000092460 UP000007755 UP000183832 UP000092553 UP000000673 UP000235965 UP000092445 UP000053097 UP000091820 UP000078541 UP000001070 UP000095300 UP000005205 UP000069940 UP000249989 UP000076502 UP000008792 UP000009192 UP000007266 UP000053825 UP000036403 UP000009046 UP000078540 UP000078200 UP000002358 UP000054359 UP000000305 UP000005203 UP000078492 UP000027135 UP000076858 UP000053105 UP000019118 UP000192223
UP000030765 UP000075880 UP000075884 UP000075886 UP000076408 UP000075883 UP000075920 UP000007062 UP000075882 UP000075840 UP000002282 UP000007801 UP000075903 UP000037069 UP000075900 UP000075902 UP000008711 UP000076407 UP000069272 UP000008744 UP000075885 UP000001819 UP000001292 UP000279307 UP000192221 UP000007798 UP000000304 UP000095301 UP000078542 UP000008237 UP000002320 UP000000311 UP000075809 UP000268350 UP000000803 UP000092443 UP000092460 UP000007755 UP000183832 UP000092553 UP000000673 UP000235965 UP000092445 UP000053097 UP000091820 UP000078541 UP000001070 UP000095300 UP000005205 UP000069940 UP000249989 UP000076502 UP000008792 UP000009192 UP000007266 UP000053825 UP000036403 UP000009046 UP000078540 UP000078200 UP000002358 UP000054359 UP000000305 UP000005203 UP000078492 UP000027135 UP000076858 UP000053105 UP000019118 UP000192223
Interpro
SUPFAM
SSF56487
SSF56487
Gene 3D
ProteinModelPortal
H9JMS0
A0A0N1PI89
A0A0N1I3I6
A0A3S2NN45
A0A2H1V7S0
A0A2A4J826
+ More
A0A212F4W5 A0A084W394 A0A182JJM3 A0A182N5M8 A0A182QRR0 A0A182YGH7 A0A182MNL6 A0A182W0I7 Q7QBA0 A0A182L7I8 A0A182I7X8 B4P8E9 B3MJ04 A0A182VAU7 A0A0L0CM11 A0A182RYI1 A0A182TK12 B3NN17 A0A182XIX9 A0A182F6N6 B4GHF6 A0A182P8X5 Q28XR1 T1PJI8 B4I7W2 A0A3L8DKP5 A0A1W4UZ01 A0A0J9U811 B4MS10 B4QGL0 A0A1I8MTI8 A0A195D5J7 E2BNK3 B0WJ73 E2AIQ3 A0A151WMF5 A0A3B0JP91 Q9W2C9 H0RNL9 A0A1Q3FPA5 A0A034WB96 A0A1A9Y7Y6 A0A1B0ARI1 F4WLF0 A0A1J1I5J6 A0A0M4E8C4 W5J9E1 Q9N9Y8 A0A2J7PI67 A0A1A9ZRG9 A0A026X3Y3 A0A1A9X3F4 A0A195FY20 B4J6A1 A0A034W761 A0A1I8PDV5 A0A158NVI9 A0A0A1WG43 A0A182H0M1 A0A154NVR5 B4LLQ2 W8C566 C5WLU2 V9IFH6 E9IWS9 B4KNS1 A0A1Y1MMJ8 D6X0V2 A0A0L7RJH8 A0A0J7L8M1 E0VBQ5 A0A0K8TZK6 A0A195BMZ9 A0A1A9VEE9 K7IM98 A0A087TCQ9 E9GHI3 A0A088ADF0 A0A195E561 A0A067R7D2 A0A0P5YL50 A0A0P5EIX5 A0A0N8AL55 A0A0P6AM69 T1E173 A0A0N0U4V5 A0A0N8E1K9 A0A147BK26 A0A0P5YQE2 A0A2H8TD94 N6TMA4 A0A0P5XQY5 A0A1W4XWS8 A0A2S2Q7B9
A0A212F4W5 A0A084W394 A0A182JJM3 A0A182N5M8 A0A182QRR0 A0A182YGH7 A0A182MNL6 A0A182W0I7 Q7QBA0 A0A182L7I8 A0A182I7X8 B4P8E9 B3MJ04 A0A182VAU7 A0A0L0CM11 A0A182RYI1 A0A182TK12 B3NN17 A0A182XIX9 A0A182F6N6 B4GHF6 A0A182P8X5 Q28XR1 T1PJI8 B4I7W2 A0A3L8DKP5 A0A1W4UZ01 A0A0J9U811 B4MS10 B4QGL0 A0A1I8MTI8 A0A195D5J7 E2BNK3 B0WJ73 E2AIQ3 A0A151WMF5 A0A3B0JP91 Q9W2C9 H0RNL9 A0A1Q3FPA5 A0A034WB96 A0A1A9Y7Y6 A0A1B0ARI1 F4WLF0 A0A1J1I5J6 A0A0M4E8C4 W5J9E1 Q9N9Y8 A0A2J7PI67 A0A1A9ZRG9 A0A026X3Y3 A0A1A9X3F4 A0A195FY20 B4J6A1 A0A034W761 A0A1I8PDV5 A0A158NVI9 A0A0A1WG43 A0A182H0M1 A0A154NVR5 B4LLQ2 W8C566 C5WLU2 V9IFH6 E9IWS9 B4KNS1 A0A1Y1MMJ8 D6X0V2 A0A0L7RJH8 A0A0J7L8M1 E0VBQ5 A0A0K8TZK6 A0A195BMZ9 A0A1A9VEE9 K7IM98 A0A087TCQ9 E9GHI3 A0A088ADF0 A0A195E561 A0A067R7D2 A0A0P5YL50 A0A0P5EIX5 A0A0N8AL55 A0A0P6AM69 T1E173 A0A0N0U4V5 A0A0N8E1K9 A0A147BK26 A0A0P5YQE2 A0A2H8TD94 N6TMA4 A0A0P5XQY5 A0A1W4XWS8 A0A2S2Q7B9
PDB
5ZE3
E-value=2.34275e-94,
Score=883
Ontologies
KEGG
GO
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.972843
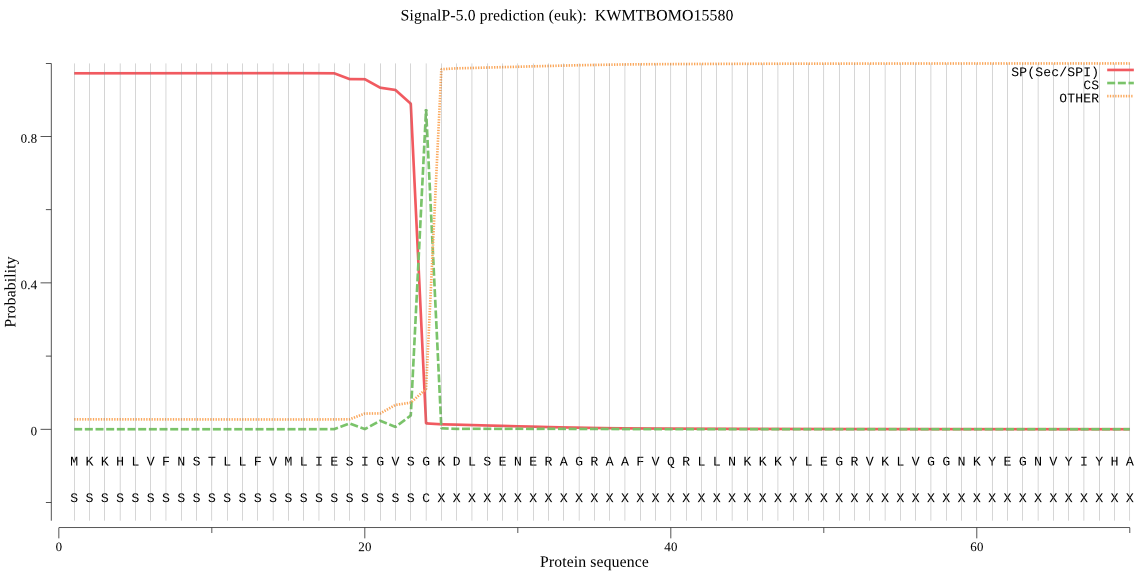
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.03118999999998
Exp number, first 60 AAs:
6.02893
Total prob of N-in:
0.32148
outside
1 - 493

Population Genetic Test Statistics
Pi
33.120753
Theta
33.420723
Tajima's D
-0.452421
CLR
0.470729
CSRT
0.252787360631968
Interpretation
Uncertain
