Gene
KWMTBOMO15576
Pre Gene Modal
BGIBMGA010728
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.963
Sequence
CDS
ATGTTTTCTAGATCCTTGACGCTAGCCGTTCTCTTTGCCGTGCTCGTGAACTTGATTGGCCAATCAATCGTCATAAATTATTTGCAGTCAATCTTGGAGATCACGCACACAAATATCAAATCGGAATTAGCTTCTGTGATTGTTGGTGTTATCCAGTTTGTAGCTGGTATCTTGGCTGCGCTTGTGACGGACAAAGTTGACCGCAAACCTTTATTAGTTGTTACCTTGGCGGGAACGGCTGCTGGTTTTACAGGACTAGGACCATTTTTCAAACTGAAAGAGCTTGGATACTCTACGGAAGGCTGGATAAATTACTTACCGTTGTTTTCGCTTGTGGCCGTTTTATTTAGCTACAGTACAGGACCCGGCTCCATTTTTCTAACCATCGTGTCTGAACTTTTCGAGGGCCCAGCAAGAGCGTCTGGTGTCACTATATCGTTCTCCGTGGCTTCTTTAACTGCGTTCCTGCTAACGAAATATTTCGCGACCTTCGTAAAACTGATTGGTCCCGTTTATTCGTATTGGTCGCTAGCCATTAATACAATTGTAGCGGGTTTATTTGTTGTAATCTTCATACCGGAAACGAAAGGAAAATCTTTTGCGGAAATCCAAAGTATACTGAAGTCGATAAACGAAGAAAATCAAAGAGATAACAGAGAGTATCAACGAACTTTTAGCTTGAAAAATACAAAACTAAAGACGCACTTTTAA
Protein
MFSRSLTLAVLFAVLVNLIGQSIVINYLQSILEITHTNIKSELASVIVGVIQFVAGILAALVTDKVDRKPLLVVTLAGTAAGFTGLGPFFKLKELGYSTEGWINYLPLFSLVAVLFSYSTGPGSIFLTIVSELFEGPARASGVTISFSVASLTAFLLTKYFATFVKLIGPVYSYWSLAINTIVAGLFVVIFIPETKGKSFAEIQSILKSINEENQRDNREYQRTFSLKNTKLKTHF
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JMH5
A0A2H1W9G7
A0A2W1BMG6
A0A0N1ICH6
A0A3S2LEG3
A0A2W1BPL4
+ More
A0A2H1VR37 A0A2A4IYC3 A0A0N0P9V6 A0A0N1PHB1 A0A0N1IJL5 A0A194QI33 A0A0N1IGH7 A0A0N0P9P7 A0A212F4M2 A0A0L7LFN4 A0A0L7KTM4 A0A0L7LC51 A0A0T6BH34 E2BLV9 A0A212FHD3 A0A1W4WJN2 A0A182GVZ2 A0A1W4W842 A0A2W1BNG5 A0A0N1PEX2 A0A1W4XAC6 Q16SU4 E9J4B0 D7EK14 A0A139WCV1 A0A1W4XJX1 D7EK13 E0W0A9 A0A195ESM4 A0A1W4W7Z0 A0A195EU19 A0A026WP92 T1PCW2 F4WP33 A0A1I8M2U2 A0A195AT95 A0A195B938 K1QZW5 A0A195EL33 A0A3L8DXN6 E2ADH2 A0A1W4WJ87 A0A0J7NVW1 A0A232F4R3 A0A158NGK6 E2B123 A0A2J7PYX6 A0A182LX60 D6X031 E2B5H9 A0A2H1VKV6 A0A151X8Q5 V5GT34 A0A1D2NCV2 A0A1Y1MWX5 A0A2H1VYV7 A0A026WQ15 A0A1W7R5M1 A0A0N0PFE6 A0A1W4WJK3 A0A0L7RBX0 A0A158NPP3 A0A0C9RGQ6 E0W2C6 A0A2P8XJ31 A0A1Y1M432 A0A3L8D924 A0A2A4JNK2 K7IP31 A0A2J7R8M9 F4W7K1 A0A154PHJ7 A0A1B0AWF5 A0A026X2K8 A0A1J1IWB4 A0A1A9XGF0 B4LPQ3 A0A195CMF0 A0A195CMH5 A0A1Y1NLV8 A0A1Y1LWD0 A0A1Y1N5Q7 A0A1D2MZ10 A0A1L8DIK0 A0A0L8GAZ6 D6X032 A0A151WM18 A0A0K8UFA2 A0A1W4X116 J9K1H3 A0A1D2MCV3 A0A1L8DIL6
A0A2H1VR37 A0A2A4IYC3 A0A0N0P9V6 A0A0N1PHB1 A0A0N1IJL5 A0A194QI33 A0A0N1IGH7 A0A0N0P9P7 A0A212F4M2 A0A0L7LFN4 A0A0L7KTM4 A0A0L7LC51 A0A0T6BH34 E2BLV9 A0A212FHD3 A0A1W4WJN2 A0A182GVZ2 A0A1W4W842 A0A2W1BNG5 A0A0N1PEX2 A0A1W4XAC6 Q16SU4 E9J4B0 D7EK14 A0A139WCV1 A0A1W4XJX1 D7EK13 E0W0A9 A0A195ESM4 A0A1W4W7Z0 A0A195EU19 A0A026WP92 T1PCW2 F4WP33 A0A1I8M2U2 A0A195AT95 A0A195B938 K1QZW5 A0A195EL33 A0A3L8DXN6 E2ADH2 A0A1W4WJ87 A0A0J7NVW1 A0A232F4R3 A0A158NGK6 E2B123 A0A2J7PYX6 A0A182LX60 D6X031 E2B5H9 A0A2H1VKV6 A0A151X8Q5 V5GT34 A0A1D2NCV2 A0A1Y1MWX5 A0A2H1VYV7 A0A026WQ15 A0A1W7R5M1 A0A0N0PFE6 A0A1W4WJK3 A0A0L7RBX0 A0A158NPP3 A0A0C9RGQ6 E0W2C6 A0A2P8XJ31 A0A1Y1M432 A0A3L8D924 A0A2A4JNK2 K7IP31 A0A2J7R8M9 F4W7K1 A0A154PHJ7 A0A1B0AWF5 A0A026X2K8 A0A1J1IWB4 A0A1A9XGF0 B4LPQ3 A0A195CMF0 A0A195CMH5 A0A1Y1NLV8 A0A1Y1LWD0 A0A1Y1N5Q7 A0A1D2MZ10 A0A1L8DIK0 A0A0L8GAZ6 D6X032 A0A151WM18 A0A0K8UFA2 A0A1W4X116 J9K1H3 A0A1D2MCV3 A0A1L8DIL6
Pubmed
EMBL
BABH01016776
ODYU01007174
SOQ49721.1
KZ150025
PZC74815.1
LADJ01026996
+ More
KPJ21155.1 RSAL01000179 RVE44945.1 PZC74816.1 ODYU01003937 SOQ43305.1 NWSH01005591 PCG64113.1 KQ459078 KPJ03808.1 KQ459986 KPJ18874.1 KPJ21147.1 KQ458761 KPJ05218.1 KPJ18875.1 KQ459256 KPJ02080.1 AGBW02010341 OWR48684.1 JTDY01001327 KOB74190.1 JTDY01006002 KOB66384.1 JTDY01001756 KOB72970.1 LJIG01000288 KRT86652.1 GL449042 EFN83325.1 AGBW02008505 OWR53139.1 JXUM01071369 JXUM01071370 JXUM01071371 JXUM01091985 KQ563956 KQ562660 KXJ73022.1 KXJ75379.1 PZC74817.1 KPJ03807.1 CH477665 EAT37544.1 GL768090 EFZ12343.1 DS497725 EFA12962.1 KQ971364 KYB25661.1 EFA12963.1 DS235858 EEB19065.1 KQ981986 KYN31228.1 KQ981965 KYN31745.1 KK107139 EZA57783.1 KA646494 AFP61123.1 GL888243 EGI63964.1 KQ976745 KYM75416.1 KQ976542 KYM81051.1 JH816771 EKC26991.1 KQ978730 KYN28965.1 QOIP01000003 RLU25240.1 GL438750 EFN68534.1 LBMM01001313 KMQ96530.1 NNAY01000987 OXU25602.1 ADTU01015114 GL444771 EFN60596.1 NEVH01020342 PNF21526.1 AXCM01000233 EFA10056.1 GL445827 EFN89065.1 ODYU01003112 SOQ41455.1 KQ982409 KYQ56668.1 GALX01003664 JAB64802.1 LJIJ01000088 ODN03069.1 GEZM01021531 JAV88955.1 ODYU01005306 SOQ46029.1 EZA57781.1 RLU25049.1 GEHC01001176 JAV46469.1 KPJ21151.1 KQ414617 KOC68300.1 ADTU01022687 GBYB01012317 JAG82084.1 DS235877 EEB19782.1 PYGN01001954 PSN32011.1 GEZM01041155 JAV80582.1 QOIP01000011 RLU16997.1 NWSH01000916 PCG73551.1 NEVH01006721 PNF37175.1 GL887844 EGI69874.1 KQ434899 KZC10964.1 JXJN01004688 KK107021 EZA62323.1 CVRI01000063 CRL04571.1 CH940648 EDW60291.1 KQ977574 KYN01893.1 KYN01895.1 GEZM01001453 JAV97755.1 GEZM01048061 JAV76640.1 GEZM01013063 JAV92788.1 LJIJ01000366 ODM98287.1 GFDF01007796 JAV06288.1 KQ422794 KOF74191.1 EFA10055.1 KQ982944 KYQ48914.1 GDHF01026962 JAI25352.1 ABLF02034162 LJIJ01001830 ODM90714.1 GFDF01007788 JAV06296.1
KPJ21155.1 RSAL01000179 RVE44945.1 PZC74816.1 ODYU01003937 SOQ43305.1 NWSH01005591 PCG64113.1 KQ459078 KPJ03808.1 KQ459986 KPJ18874.1 KPJ21147.1 KQ458761 KPJ05218.1 KPJ18875.1 KQ459256 KPJ02080.1 AGBW02010341 OWR48684.1 JTDY01001327 KOB74190.1 JTDY01006002 KOB66384.1 JTDY01001756 KOB72970.1 LJIG01000288 KRT86652.1 GL449042 EFN83325.1 AGBW02008505 OWR53139.1 JXUM01071369 JXUM01071370 JXUM01071371 JXUM01091985 KQ563956 KQ562660 KXJ73022.1 KXJ75379.1 PZC74817.1 KPJ03807.1 CH477665 EAT37544.1 GL768090 EFZ12343.1 DS497725 EFA12962.1 KQ971364 KYB25661.1 EFA12963.1 DS235858 EEB19065.1 KQ981986 KYN31228.1 KQ981965 KYN31745.1 KK107139 EZA57783.1 KA646494 AFP61123.1 GL888243 EGI63964.1 KQ976745 KYM75416.1 KQ976542 KYM81051.1 JH816771 EKC26991.1 KQ978730 KYN28965.1 QOIP01000003 RLU25240.1 GL438750 EFN68534.1 LBMM01001313 KMQ96530.1 NNAY01000987 OXU25602.1 ADTU01015114 GL444771 EFN60596.1 NEVH01020342 PNF21526.1 AXCM01000233 EFA10056.1 GL445827 EFN89065.1 ODYU01003112 SOQ41455.1 KQ982409 KYQ56668.1 GALX01003664 JAB64802.1 LJIJ01000088 ODN03069.1 GEZM01021531 JAV88955.1 ODYU01005306 SOQ46029.1 EZA57781.1 RLU25049.1 GEHC01001176 JAV46469.1 KPJ21151.1 KQ414617 KOC68300.1 ADTU01022687 GBYB01012317 JAG82084.1 DS235877 EEB19782.1 PYGN01001954 PSN32011.1 GEZM01041155 JAV80582.1 QOIP01000011 RLU16997.1 NWSH01000916 PCG73551.1 NEVH01006721 PNF37175.1 GL887844 EGI69874.1 KQ434899 KZC10964.1 JXJN01004688 KK107021 EZA62323.1 CVRI01000063 CRL04571.1 CH940648 EDW60291.1 KQ977574 KYN01893.1 KYN01895.1 GEZM01001453 JAV97755.1 GEZM01048061 JAV76640.1 GEZM01013063 JAV92788.1 LJIJ01000366 ODM98287.1 GFDF01007796 JAV06288.1 KQ422794 KOF74191.1 EFA10055.1 KQ982944 KYQ48914.1 GDHF01026962 JAI25352.1 ABLF02034162 LJIJ01001830 ODM90714.1 GFDF01007788 JAV06296.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000218220
UP000053268
UP000007151
+ More
UP000037510 UP000008237 UP000192223 UP000069940 UP000249989 UP000008820 UP000007266 UP000009046 UP000078541 UP000053097 UP000007755 UP000095301 UP000078540 UP000005408 UP000078492 UP000279307 UP000000311 UP000036403 UP000215335 UP000005205 UP000235965 UP000075883 UP000075809 UP000094527 UP000053825 UP000245037 UP000002358 UP000076502 UP000092460 UP000183832 UP000092443 UP000008792 UP000078542 UP000053454 UP000007819
UP000037510 UP000008237 UP000192223 UP000069940 UP000249989 UP000008820 UP000007266 UP000009046 UP000078541 UP000053097 UP000007755 UP000095301 UP000078540 UP000005408 UP000078492 UP000279307 UP000000311 UP000036403 UP000215335 UP000005205 UP000235965 UP000075883 UP000075809 UP000094527 UP000053825 UP000245037 UP000002358 UP000076502 UP000092460 UP000183832 UP000092443 UP000008792 UP000078542 UP000053454 UP000007819
PRIDE
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JMH5
A0A2H1W9G7
A0A2W1BMG6
A0A0N1ICH6
A0A3S2LEG3
A0A2W1BPL4
+ More
A0A2H1VR37 A0A2A4IYC3 A0A0N0P9V6 A0A0N1PHB1 A0A0N1IJL5 A0A194QI33 A0A0N1IGH7 A0A0N0P9P7 A0A212F4M2 A0A0L7LFN4 A0A0L7KTM4 A0A0L7LC51 A0A0T6BH34 E2BLV9 A0A212FHD3 A0A1W4WJN2 A0A182GVZ2 A0A1W4W842 A0A2W1BNG5 A0A0N1PEX2 A0A1W4XAC6 Q16SU4 E9J4B0 D7EK14 A0A139WCV1 A0A1W4XJX1 D7EK13 E0W0A9 A0A195ESM4 A0A1W4W7Z0 A0A195EU19 A0A026WP92 T1PCW2 F4WP33 A0A1I8M2U2 A0A195AT95 A0A195B938 K1QZW5 A0A195EL33 A0A3L8DXN6 E2ADH2 A0A1W4WJ87 A0A0J7NVW1 A0A232F4R3 A0A158NGK6 E2B123 A0A2J7PYX6 A0A182LX60 D6X031 E2B5H9 A0A2H1VKV6 A0A151X8Q5 V5GT34 A0A1D2NCV2 A0A1Y1MWX5 A0A2H1VYV7 A0A026WQ15 A0A1W7R5M1 A0A0N0PFE6 A0A1W4WJK3 A0A0L7RBX0 A0A158NPP3 A0A0C9RGQ6 E0W2C6 A0A2P8XJ31 A0A1Y1M432 A0A3L8D924 A0A2A4JNK2 K7IP31 A0A2J7R8M9 F4W7K1 A0A154PHJ7 A0A1B0AWF5 A0A026X2K8 A0A1J1IWB4 A0A1A9XGF0 B4LPQ3 A0A195CMF0 A0A195CMH5 A0A1Y1NLV8 A0A1Y1LWD0 A0A1Y1N5Q7 A0A1D2MZ10 A0A1L8DIK0 A0A0L8GAZ6 D6X032 A0A151WM18 A0A0K8UFA2 A0A1W4X116 J9K1H3 A0A1D2MCV3 A0A1L8DIL6
A0A2H1VR37 A0A2A4IYC3 A0A0N0P9V6 A0A0N1PHB1 A0A0N1IJL5 A0A194QI33 A0A0N1IGH7 A0A0N0P9P7 A0A212F4M2 A0A0L7LFN4 A0A0L7KTM4 A0A0L7LC51 A0A0T6BH34 E2BLV9 A0A212FHD3 A0A1W4WJN2 A0A182GVZ2 A0A1W4W842 A0A2W1BNG5 A0A0N1PEX2 A0A1W4XAC6 Q16SU4 E9J4B0 D7EK14 A0A139WCV1 A0A1W4XJX1 D7EK13 E0W0A9 A0A195ESM4 A0A1W4W7Z0 A0A195EU19 A0A026WP92 T1PCW2 F4WP33 A0A1I8M2U2 A0A195AT95 A0A195B938 K1QZW5 A0A195EL33 A0A3L8DXN6 E2ADH2 A0A1W4WJ87 A0A0J7NVW1 A0A232F4R3 A0A158NGK6 E2B123 A0A2J7PYX6 A0A182LX60 D6X031 E2B5H9 A0A2H1VKV6 A0A151X8Q5 V5GT34 A0A1D2NCV2 A0A1Y1MWX5 A0A2H1VYV7 A0A026WQ15 A0A1W7R5M1 A0A0N0PFE6 A0A1W4WJK3 A0A0L7RBX0 A0A158NPP3 A0A0C9RGQ6 E0W2C6 A0A2P8XJ31 A0A1Y1M432 A0A3L8D924 A0A2A4JNK2 K7IP31 A0A2J7R8M9 F4W7K1 A0A154PHJ7 A0A1B0AWF5 A0A026X2K8 A0A1J1IWB4 A0A1A9XGF0 B4LPQ3 A0A195CMF0 A0A195CMH5 A0A1Y1NLV8 A0A1Y1LWD0 A0A1Y1N5Q7 A0A1D2MZ10 A0A1L8DIK0 A0A0L8GAZ6 D6X032 A0A151WM18 A0A0K8UFA2 A0A1W4X116 J9K1H3 A0A1D2MCV3 A0A1L8DIL6
PDB
4LDS
E-value=2.52075e-12,
Score=171
Ontologies
GO
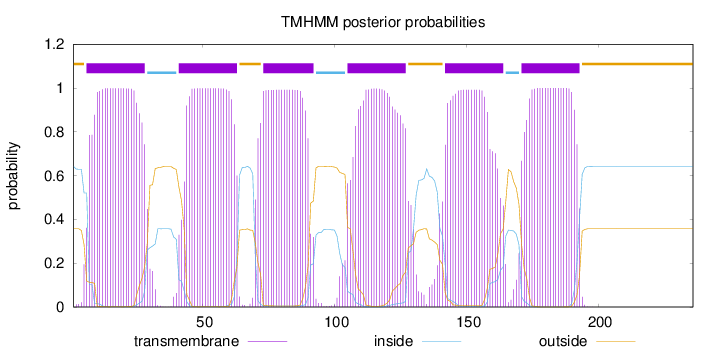
Topology
Length:
236
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
129.68265
Exp number, first 60 AAs:
40.51112
Total prob of N-in:
0.64233
POSSIBLE N-term signal
sequence
outside
1 - 5
TMhelix
6 - 28
inside
29 - 40
TMhelix
41 - 63
outside
64 - 72
TMhelix
73 - 92
inside
93 - 104
TMhelix
105 - 127
outside
128 - 141
TMhelix
142 - 164
inside
165 - 170
TMhelix
171 - 193
outside
194 - 236
Population Genetic Test Statistics
Pi
178.215636
Theta
140.59244
Tajima's D
0.745096
CLR
0
CSRT
0.589970501474926
Interpretation
Uncertain
