Gene
KWMTBOMO15571
Pre Gene Modal
BGIBMGA010730
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Full name
Facilitated trehalose transporter Tret1-2 homolog
+ More
Facilitated trehalose transporter Tret1
Facilitated trehalose transporter Tret1
Location in the cell
PlasmaMembrane Reliability : 4.973
Sequence
CDS
ATGTCGGAGAGATTACGTTGTTTGATCGGCTTCTGTTTGGGTCCGTACCTCTCCTACCTCCAGTTTGGATTCGTGCCTATAATTTTAATAATAATATTCATTGCGCTCAGCTTCTTCTTGAACGAATCTCCAGTGTATTTTGTTATTAAGGGCGACGAAGAATCAGCCAAAAAGGCTCTAAATTACTTTGGTCGCAGCAAAGATGTAAATTACGAACTGAAAATTATGCTTCAAAATGCAAAAGAGAACGCACAAGGAAAAAGGAAATGGCTTGAACTGTTTCAGAGGAGTAACAGAAGAGCACTCCTGATTATTATTCCATTATATATAATCGAACAAGCAAGCGGTGTTATAGCTGTTATATTAAATGCAACAGCAATTTTCAAATTAGCCGGCTCATCAGTTGAACCGGCAGTTGCGACCATGATAGTTGGCGTTACTAATCTGACGGGAAGCTTAATCTCGCCATTTGTCGTGGATAGGTATGGAAGGAAGGTGCTTTTGTTGTTTTCAGCAGGAGGGTGCTGTCTGGCAATGACTGCTTTAGGAACTTATTTTTATTTTCGACAAATGTTTCCGGAGTCTGTTGGATCCCTGGGCTGGTTACCGCTTACTGCTTTAGTTGCAGCTCTGTTTACGTACAGCGTTGGTCTGACAATAATACCACACGCTCTAGCAGGGGAAATGTTTAGTCCGAAAATGCGTGGGCTAGGCACCTCTCTAGGTTTGACAGTCGCCAATGGCTCCGGTCTGATAACCACCAGCATTTATTCTTACCTAGCAGTAAACGTCGGCATCCATTACGCTTTCTGGATGTTCGCTGTCTTTGACTTGGTTGGCGTCATCTTCACCATCTATGTCGTGCCGGAGACCAAAGGAAAAAGCTTATCTGACATTAACGCTATGCTAGCAAAATAA
Protein
MSERLRCLIGFCLGPYLSYLQFGFVPIILIIIFIALSFFLNESPVYFVIKGDEESAKKALNYFGRSKDVNYELKIMLQNAKENAQGKRKWLELFQRSNRRALLIIIPLYIIEQASGVIAVILNATAIFKLAGSSVEPAVATMIVGVTNLTGSLISPFVVDRYGRKVLLLFSAGGCCLAMTALGTYFYFRQMFPESVGSLGWLPLTALVAALFTYSVGLTIIPHALAGEMFSPKMRGLGTSLGLTVANGSGLITTSIYSYLAVNVGIHYAFWMFAVFDLVGVIFTIYVVPETKGKSLSDINAMLAK
Summary
Description
Fails to transport trehalose.
High-capacity facilitative transporter for trehalose, required to induce anhydrobiosis. Anhydrobiotic larvae can survive almost complete dehydration. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph.
High-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph.
High-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph (By similarity).
Low-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph (By similarity).
High-capacity facilitative transporter for trehalose, required to induce anhydrobiosis. Anhydrobiotic larvae can survive almost complete dehydration. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph.
High-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph.
High-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph (By similarity).
Low-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph (By similarity).
Biophysicochemical Properties
114.5 mM for trehalose
45.74 mM for trehalose
45.74 mM for trehalose
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Trehalose transporter subfamily.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Trehalose transporter subfamily.
Keywords
Cell membrane
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Stress response
Sugar transport
Transport
Alternative splicing
Phosphoprotein
Feature
chain Facilitated trehalose transporter Tret1-2 homolog
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A0N0PFE7
A0A212FHD3
A0A0N0PFE6
A0A0N1PEX2
A0A212ELA9
A0A0L7LF84
+ More
A0A2H1VYV7 A0A2W1BNG5 A0A2P8YD54 A0A0N0PFQ6 A0A1L8DFA9 A0A0J9RBH4 B4QBN3 A0A1L8DF29 A0A067RCA0 A0A182T6H7 A0A2H1V3K1 Q16SU5 A0A1L8DFC4 A0A2M4A4V5 A0A194R1W3 A0A2A4JZZ4 A0A194PW18 A0A2P8XWS8 A0A2H1WAI1 A0A1I8MXU6 A0A182NLP8 A0A2A4J788 A0A0P9BY82 W5JNP3 B4HNS1 A0A182GL69 A0A182GVZ1 A0A0R1DWD3 A0A182K5N6 A5LGM7 A0A084VDW1 A0A2M3YZE1 A0A2M4A3H1 A0A182T9M2 A0A182MM04 A0A2M4A537 T1DE67 T1DKQ7 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182FCI5 A0A182TSI6 A0A2M4BMJ9 Q8MKK4 A0A2M4A4U2 A0A1B0C869 D4AHX3 A0A182VIR9 A0A182J8U1 A0A194PK39 A0A2C9GQB0 A0A2M4BKT8 A0A1L8DF37 A0A1L8DF53 A0A182Y131 Q7PIR5-2 A0A182HLX0 A0A182TY80 A0A182X524 A0A182S579 S4P7H5 T1DG93 A0A1W4VCC4 A0A2M3ZF95 A0A336M0J9 A0A182RMH4 E0W471 A0A182MWR6 K7IP31 A0A182WKF0 A0A182GLX7 A0A182YLM9 A0A2M3ZFG5 A0A0R3NRR1 A0A2M4CVC5 W5J543 A0A1B0C868 U5EQL0 B0WC46 B3NSE1 A0A182QT48 A0A0Q9WFV5 A0A182VS58 B3MG58 A0A182HJZ0 A0A0K8W3N6 A0A182Q863 B4P624 A0A1Y9H2R2 W8B3Q4 A0A1L8DIL5 A0A2H1WAF6
A0A2H1VYV7 A0A2W1BNG5 A0A2P8YD54 A0A0N0PFQ6 A0A1L8DFA9 A0A0J9RBH4 B4QBN3 A0A1L8DF29 A0A067RCA0 A0A182T6H7 A0A2H1V3K1 Q16SU5 A0A1L8DFC4 A0A2M4A4V5 A0A194R1W3 A0A2A4JZZ4 A0A194PW18 A0A2P8XWS8 A0A2H1WAI1 A0A1I8MXU6 A0A182NLP8 A0A2A4J788 A0A0P9BY82 W5JNP3 B4HNS1 A0A182GL69 A0A182GVZ1 A0A0R1DWD3 A0A182K5N6 A5LGM7 A0A084VDW1 A0A2M3YZE1 A0A2M4A3H1 A0A182T9M2 A0A182MM04 A0A2M4A537 T1DE67 T1DKQ7 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182FCI5 A0A182TSI6 A0A2M4BMJ9 Q8MKK4 A0A2M4A4U2 A0A1B0C869 D4AHX3 A0A182VIR9 A0A182J8U1 A0A194PK39 A0A2C9GQB0 A0A2M4BKT8 A0A1L8DF37 A0A1L8DF53 A0A182Y131 Q7PIR5-2 A0A182HLX0 A0A182TY80 A0A182X524 A0A182S579 S4P7H5 T1DG93 A0A1W4VCC4 A0A2M3ZF95 A0A336M0J9 A0A182RMH4 E0W471 A0A182MWR6 K7IP31 A0A182WKF0 A0A182GLX7 A0A182YLM9 A0A2M3ZFG5 A0A0R3NRR1 A0A2M4CVC5 W5J543 A0A1B0C868 U5EQL0 B0WC46 B3NSE1 A0A182QT48 A0A0Q9WFV5 A0A182VS58 B3MG58 A0A182HJZ0 A0A0K8W3N6 A0A182Q863 B4P624 A0A1Y9H2R2 W8B3Q4 A0A1L8DIL5 A0A2H1WAF6
Pubmed
EMBL
LADJ01026996
KPJ21154.1
AGBW02008505
OWR53139.1
KPJ21151.1
KQ459078
+ More
KPJ03807.1 AGBW02014089 OWR42276.1 JTDY01001327 KOB74188.1 ODYU01005306 SOQ46029.1 KZ150025 PZC74817.1 PYGN01000690 PSN42188.1 KPJ21152.1 GFDF01009044 JAV05040.1 CM002911 KMY93024.1 CM000362 EDX06650.1 GFDF01009043 JAV05041.1 KK852731 KDR17493.1 ODYU01000519 SOQ35423.1 CH477665 EAT37543.1 GFDF01009007 JAV05077.1 GGFK01002478 MBW35799.1 KQ460870 KPJ11682.1 NWSH01000363 PCG76972.1 KQ459589 KPI97522.1 PYGN01001230 PSN36458.1 ODYU01007368 SOQ50068.1 NWSH01002911 PCG67310.1 CH902619 KPU76395.1 ADMH02000542 ETN65997.1 CH480816 EDW47436.1 JXUM01071368 JXUM01071369 KQ562660 KXJ75378.1 JXUM01091981 JXUM01091982 KQ563956 KXJ73021.1 CM000158 KRJ99505.1 AB272983 BAF63703.1 ATLV01011809 KE524723 KFB36155.1 GGFM01000865 MBW21616.1 GGFK01002032 MBW35353.1 AXCM01000233 GGFK01002603 MBW35924.1 GALA01001167 JAA93685.1 GAMD01000836 JAB00755.1 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 GGFJ01004857 MBW53998.1 AB369551 AE013599 AY122079 AAM52591.1 AAM68715.1 BAF96745.1 GGFK01002450 MBW35771.1 AJWK01000224 AJWK01000225 AB550001 BAI83422.1 KQ459601 KPI93687.1 APCN01004269 GGFJ01004545 MBW53686.1 GFDF01009012 JAV05072.1 GFDF01009013 JAV05071.1 AAAB01008960 AB369548 BAF96742.1 EAA44045.3 GAIX01004609 JAA87951.1 GAMD01002846 JAA98744.1 GGFM01006410 MBW27161.1 UFQT01000389 SSX23856.1 DS235886 EEB20427.1 AXCM01005103 JXUM01001466 KQ560121 KXJ84364.1 GGFM01006409 MBW27160.1 CM000071 KRT01871.1 GGFL01004973 MBW69151.1 ADMH02002130 ETN58528.1 GANO01004191 JAB55680.1 DS231885 EDS43198.1 CH954179 EDV56443.1 AXCN02000090 CH940648 KRF79918.1 EDV36753.1 APCN01002224 GDHF01014137 GDHF01006620 JAI38177.1 JAI45694.1 AXCN02000985 EDW90899.1 GAMC01018654 JAB87901.1 GFDF01007792 JAV06292.1 SOQ50069.1
KPJ03807.1 AGBW02014089 OWR42276.1 JTDY01001327 KOB74188.1 ODYU01005306 SOQ46029.1 KZ150025 PZC74817.1 PYGN01000690 PSN42188.1 KPJ21152.1 GFDF01009044 JAV05040.1 CM002911 KMY93024.1 CM000362 EDX06650.1 GFDF01009043 JAV05041.1 KK852731 KDR17493.1 ODYU01000519 SOQ35423.1 CH477665 EAT37543.1 GFDF01009007 JAV05077.1 GGFK01002478 MBW35799.1 KQ460870 KPJ11682.1 NWSH01000363 PCG76972.1 KQ459589 KPI97522.1 PYGN01001230 PSN36458.1 ODYU01007368 SOQ50068.1 NWSH01002911 PCG67310.1 CH902619 KPU76395.1 ADMH02000542 ETN65997.1 CH480816 EDW47436.1 JXUM01071368 JXUM01071369 KQ562660 KXJ75378.1 JXUM01091981 JXUM01091982 KQ563956 KXJ73021.1 CM000158 KRJ99505.1 AB272983 BAF63703.1 ATLV01011809 KE524723 KFB36155.1 GGFM01000865 MBW21616.1 GGFK01002032 MBW35353.1 AXCM01000233 GGFK01002603 MBW35924.1 GALA01001167 JAA93685.1 GAMD01000836 JAB00755.1 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 GGFJ01004857 MBW53998.1 AB369551 AE013599 AY122079 AAM52591.1 AAM68715.1 BAF96745.1 GGFK01002450 MBW35771.1 AJWK01000224 AJWK01000225 AB550001 BAI83422.1 KQ459601 KPI93687.1 APCN01004269 GGFJ01004545 MBW53686.1 GFDF01009012 JAV05072.1 GFDF01009013 JAV05071.1 AAAB01008960 AB369548 BAF96742.1 EAA44045.3 GAIX01004609 JAA87951.1 GAMD01002846 JAA98744.1 GGFM01006410 MBW27161.1 UFQT01000389 SSX23856.1 DS235886 EEB20427.1 AXCM01005103 JXUM01001466 KQ560121 KXJ84364.1 GGFM01006409 MBW27160.1 CM000071 KRT01871.1 GGFL01004973 MBW69151.1 ADMH02002130 ETN58528.1 GANO01004191 JAB55680.1 DS231885 EDS43198.1 CH954179 EDV56443.1 AXCN02000090 CH940648 KRF79918.1 EDV36753.1 APCN01002224 GDHF01014137 GDHF01006620 JAI38177.1 JAI45694.1 AXCN02000985 EDW90899.1 GAMC01018654 JAB87901.1 GFDF01007792 JAV06292.1 SOQ50069.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000037510
UP000245037
UP000000304
+ More
UP000027135 UP000075901 UP000008820 UP000218220 UP000095301 UP000075884 UP000007801 UP000000673 UP000001292 UP000069940 UP000249989 UP000002282 UP000075881 UP000030765 UP000075883 UP000075882 UP000007062 UP000076407 UP000069272 UP000075902 UP000000803 UP000092461 UP000075903 UP000075880 UP000075840 UP000076408 UP000075900 UP000192221 UP000009046 UP000002358 UP000075920 UP000001819 UP000002320 UP000008711 UP000075886 UP000008792
UP000027135 UP000075901 UP000008820 UP000218220 UP000095301 UP000075884 UP000007801 UP000000673 UP000001292 UP000069940 UP000249989 UP000002282 UP000075881 UP000030765 UP000075883 UP000075882 UP000007062 UP000076407 UP000069272 UP000075902 UP000000803 UP000092461 UP000075903 UP000075880 UP000075840 UP000076408 UP000075900 UP000192221 UP000009046 UP000002358 UP000075920 UP000001819 UP000002320 UP000008711 UP000075886 UP000008792
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A0N0PFE7
A0A212FHD3
A0A0N0PFE6
A0A0N1PEX2
A0A212ELA9
A0A0L7LF84
+ More
A0A2H1VYV7 A0A2W1BNG5 A0A2P8YD54 A0A0N0PFQ6 A0A1L8DFA9 A0A0J9RBH4 B4QBN3 A0A1L8DF29 A0A067RCA0 A0A182T6H7 A0A2H1V3K1 Q16SU5 A0A1L8DFC4 A0A2M4A4V5 A0A194R1W3 A0A2A4JZZ4 A0A194PW18 A0A2P8XWS8 A0A2H1WAI1 A0A1I8MXU6 A0A182NLP8 A0A2A4J788 A0A0P9BY82 W5JNP3 B4HNS1 A0A182GL69 A0A182GVZ1 A0A0R1DWD3 A0A182K5N6 A5LGM7 A0A084VDW1 A0A2M3YZE1 A0A2M4A3H1 A0A182T9M2 A0A182MM04 A0A2M4A537 T1DE67 T1DKQ7 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182FCI5 A0A182TSI6 A0A2M4BMJ9 Q8MKK4 A0A2M4A4U2 A0A1B0C869 D4AHX3 A0A182VIR9 A0A182J8U1 A0A194PK39 A0A2C9GQB0 A0A2M4BKT8 A0A1L8DF37 A0A1L8DF53 A0A182Y131 Q7PIR5-2 A0A182HLX0 A0A182TY80 A0A182X524 A0A182S579 S4P7H5 T1DG93 A0A1W4VCC4 A0A2M3ZF95 A0A336M0J9 A0A182RMH4 E0W471 A0A182MWR6 K7IP31 A0A182WKF0 A0A182GLX7 A0A182YLM9 A0A2M3ZFG5 A0A0R3NRR1 A0A2M4CVC5 W5J543 A0A1B0C868 U5EQL0 B0WC46 B3NSE1 A0A182QT48 A0A0Q9WFV5 A0A182VS58 B3MG58 A0A182HJZ0 A0A0K8W3N6 A0A182Q863 B4P624 A0A1Y9H2R2 W8B3Q4 A0A1L8DIL5 A0A2H1WAF6
A0A2H1VYV7 A0A2W1BNG5 A0A2P8YD54 A0A0N0PFQ6 A0A1L8DFA9 A0A0J9RBH4 B4QBN3 A0A1L8DF29 A0A067RCA0 A0A182T6H7 A0A2H1V3K1 Q16SU5 A0A1L8DFC4 A0A2M4A4V5 A0A194R1W3 A0A2A4JZZ4 A0A194PW18 A0A2P8XWS8 A0A2H1WAI1 A0A1I8MXU6 A0A182NLP8 A0A2A4J788 A0A0P9BY82 W5JNP3 B4HNS1 A0A182GL69 A0A182GVZ1 A0A0R1DWD3 A0A182K5N6 A5LGM7 A0A084VDW1 A0A2M3YZE1 A0A2M4A3H1 A0A182T9M2 A0A182MM04 A0A2M4A537 T1DE67 T1DKQ7 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182FCI5 A0A182TSI6 A0A2M4BMJ9 Q8MKK4 A0A2M4A4U2 A0A1B0C869 D4AHX3 A0A182VIR9 A0A182J8U1 A0A194PK39 A0A2C9GQB0 A0A2M4BKT8 A0A1L8DF37 A0A1L8DF53 A0A182Y131 Q7PIR5-2 A0A182HLX0 A0A182TY80 A0A182X524 A0A182S579 S4P7H5 T1DG93 A0A1W4VCC4 A0A2M3ZF95 A0A336M0J9 A0A182RMH4 E0W471 A0A182MWR6 K7IP31 A0A182WKF0 A0A182GLX7 A0A182YLM9 A0A2M3ZFG5 A0A0R3NRR1 A0A2M4CVC5 W5J543 A0A1B0C868 U5EQL0 B0WC46 B3NSE1 A0A182QT48 A0A0Q9WFV5 A0A182VS58 B3MG58 A0A182HJZ0 A0A0K8W3N6 A0A182Q863 B4P624 A0A1Y9H2R2 W8B3Q4 A0A1L8DIL5 A0A2H1WAF6
PDB
4YBQ
E-value=1.56893e-19,
Score=235
Ontologies
GO
Topology
Subcellular location
Cell membrane
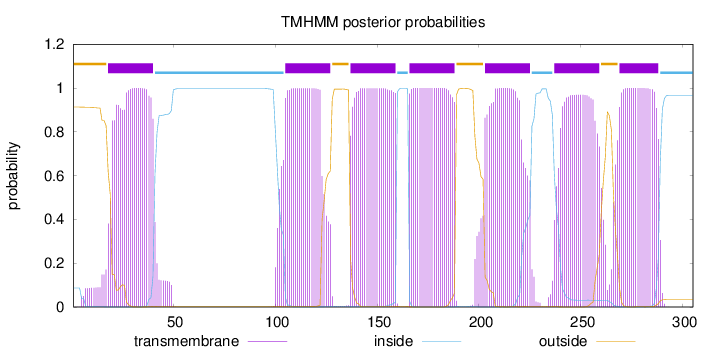
Length:
305
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
158.05241
Exp number, first 60 AAs:
23.52985
Total prob of N-in:
0.08685
POSSIBLE N-term signal
sequence
outside
1 - 17
TMhelix
18 - 40
inside
41 - 104
TMhelix
105 - 127
outside
128 - 136
TMhelix
137 - 159
inside
160 - 165
TMhelix
166 - 188
outside
189 - 202
TMhelix
203 - 225
inside
226 - 236
TMhelix
237 - 259
outside
260 - 268
TMhelix
269 - 288
inside
289 - 305
Population Genetic Test Statistics
Pi
196.740787
Theta
167.292623
Tajima's D
0.598323
CLR
0.140831
CSRT
0.541772911354432
Interpretation
Uncertain
