Gene
KWMTBOMO15565
Annotation
PREDICTED:_attractin-like_protein_1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 0.989
Sequence
CDS
ATGCACGGTTCAGTAGTTATAACGGAACATCCGTATAAAGTGTCGACCGGCGTGTGGCAGAGCGTCCCGACGCACGGGGACGCGCCCGCCGCCCGCTCCGCGCACTCCGCCGTGCTGTACGGGCACGAGCTCATCGTGTACGGGGGCGCCGTGGCCAGCGACGACCCGGAGCGGGGTGGGGGGTTGGCAGGATTGGAGGGGAGAGCTGGCCGGCTGAGCAACGAGGTGTGGCGCGCGCGGCTGGACACCGAGCCCGTGCGCTGGCACAACGCCACGCCCGCCGCCTGCTCGCCGCACAGGCACGCGCCGCTCGCGCACTGCGGAGGTCTCCACGTGTCCGGTCACACAGCGGTTCTGGTATACTTAGGAGCCACCAAAAAGCCCGTGATGCTCGTCTTCTTCGGGCATTCGGACATATACGGATATTTACATATCGTTCAGGAGTACTACATAGAAGAGAACGTGTGGTCGGCGGCCCCCACGCAGGGCTGGCCGGCCCGCGCGGGGTTCGGTCACTCGGCCGCGTGGGACGCGCTCTCCAGGCGAGTCTACATACACGGCGGGCTCGTCTCGGAGTCGGAAGCTACTCAGGCGCCGTCCGCAGCGTTATTTGAATACGATGTGGAGTCCAGGGTGTTCAAACCGCTGCCTTCCGCTCCCACACCTAGATATTTGCACTCGGCGGTTTTCGTATCACCAGGCGTGATGTTGGTGTTCGGCGGTAACGCCCACAACGACAGTGCCGCGGCCGCCATCACCAACCCGACCGGCGCGCGCTGCTACTCGGCCGGCGCCCTGCTCTACGACGTCAGATGTTCCCGCTGGTGGGAGACGCGCAGCCCGGCGGGGGGCGCGCGGGCGGCGGGGGCGGCGTGCGCGCTGCCACAGCACTCGCGCCCGGCCGCGCTGCTGCACGGAGGTTTCAACGGGCGCCTGCGCTCGGACGCGCTGGTGTACGAGGCGGGCGAGCGGTGCGCGGCGCATGCGCACGAGGACGCCTGCCTCAACAGCACGCGCCGCGCGGCCGTGGCGTGCGCGTGGCGGGAGGACCTGCAGCTGTGCGTGCAACTAGAAGAGATAGGCTGGAAGGAGTCTTTCGACGATGCCGTGAGGGCCTGCGCCATCGAACCGACTTTTGGTAAGCTTGTATACTTTATTGTATCTTTATTTTGA
Protein
MHGSVVITEHPYKVSTGVWQSVPTHGDAPAARSAHSAVLYGHELIVYGGAVASDDPERGGGLAGLEGRAGRLSNEVWRARLDTEPVRWHNATPAACSPHRHAPLAHCGGLHVSGHTAVLVYLGATKKPVMLVFFGHSDIYGYLHIVQEYYIEENVWSAAPTQGWPARAGFGHSAAWDALSRRVYIHGGLVSESEATQAPSAALFEYDVESRVFKPLPSAPTPRYLHSAVFVSPGVMLVFGGNAHNDSAAAAITNPTGARCYSAGALLYDVRCSRWWETRSPAGGARAAGAACALPQHSRPAALLHGGFNGRLRSDALVYEAGERCAAHAHEDACLNSTRRAAVACAWREDLQLCVQLEEIGWKESFDDAVRACAIEPTFGKLVYFIVSLF
Summary
Uniprot
H9JMR3
A0A2H1W201
A0A2W1BKK8
A0A194QJX6
A0A3S2LVK9
A0A1Y1KZX7
+ More
A0A154PI53 A0A0M4ET48 B4JRY4 V5FYV1 B4M631 A0A026X3D0 A0A1B0FKS7 A0A1A9YEX4 A0A1B0BSW5 A0A1B0A3R6 A0A0Q9X2N6 A0A1A9VGB4 A0A1A9WT77 B4KBA1 A0A088AMS9 A0A3L8DKN8 D6X0Y8 A0A0L7RJI8 E2BYN7 A0A2R7VX70 A0A1W4X0H6 A0A1W4XB92 A0A3B0K6S3 A0A0T6AW74 E2AUP6 F4WJL9 A0A1W4X0E8 A0A3B0JYV9 A0A3B0JK42 A0A0J7KRT9 A0A2A3EDI7 A0A195BEV8 A0A3B0K3X1 A0A224XIR3 A0A195EEX6 A0A151WEJ9 A0A2J7Q5V8 Q29BH4 B4GNY4 A0A195FGN3 A0A0K8USS7 A0A034VWX1 A0A1I8PS18 A0A2J7Q5V6 A0A2J7Q5W5 A0A034VYJ1 A0A2J7Q5W1 T1HMB1 Q6NP06 A0A2J7Q5V1 A0A2J7Q5V2 E9IAJ2 A0A2J7Q5W2 A0A0A1WFU4 A0A0K8VUP9 Q960R8 A0A1Q3F5K1 A0A0K8V1S0 A0A1W4UQ01 B4PRC5 Q9VB20 B6IDV0 A0A1Q3F5F7 A0A1Q3F5J0 A0A3Q0JGC0 A0A0L0CJ26 B3P5Z8 A0A2P8XWR9 B3LXJ0 A0A1B6D320 A0A023F3J4 W8B6R9 B4NAQ5 A0A182JY31 A0A1I8N2W6 T1PBU6 A0A0M8ZSK6 A0A182H8U6 Q172L4 Q172L3 A0A182LX91 A0A182R891 A0A182YLM0 A0A182VS49 B0WJT8 A0A0A9YNY9 A0A182JLU8 A0A0A9YIC6 A0A182TLW8 A0A0A9YTC1 A0A0A9YMG1 A0A0A9YMG6
A0A154PI53 A0A0M4ET48 B4JRY4 V5FYV1 B4M631 A0A026X3D0 A0A1B0FKS7 A0A1A9YEX4 A0A1B0BSW5 A0A1B0A3R6 A0A0Q9X2N6 A0A1A9VGB4 A0A1A9WT77 B4KBA1 A0A088AMS9 A0A3L8DKN8 D6X0Y8 A0A0L7RJI8 E2BYN7 A0A2R7VX70 A0A1W4X0H6 A0A1W4XB92 A0A3B0K6S3 A0A0T6AW74 E2AUP6 F4WJL9 A0A1W4X0E8 A0A3B0JYV9 A0A3B0JK42 A0A0J7KRT9 A0A2A3EDI7 A0A195BEV8 A0A3B0K3X1 A0A224XIR3 A0A195EEX6 A0A151WEJ9 A0A2J7Q5V8 Q29BH4 B4GNY4 A0A195FGN3 A0A0K8USS7 A0A034VWX1 A0A1I8PS18 A0A2J7Q5V6 A0A2J7Q5W5 A0A034VYJ1 A0A2J7Q5W1 T1HMB1 Q6NP06 A0A2J7Q5V1 A0A2J7Q5V2 E9IAJ2 A0A2J7Q5W2 A0A0A1WFU4 A0A0K8VUP9 Q960R8 A0A1Q3F5K1 A0A0K8V1S0 A0A1W4UQ01 B4PRC5 Q9VB20 B6IDV0 A0A1Q3F5F7 A0A1Q3F5J0 A0A3Q0JGC0 A0A0L0CJ26 B3P5Z8 A0A2P8XWR9 B3LXJ0 A0A1B6D320 A0A023F3J4 W8B6R9 B4NAQ5 A0A182JY31 A0A1I8N2W6 T1PBU6 A0A0M8ZSK6 A0A182H8U6 Q172L4 Q172L3 A0A182LX91 A0A182R891 A0A182YLM0 A0A182VS49 B0WJT8 A0A0A9YNY9 A0A182JLU8 A0A0A9YIC6 A0A182TLW8 A0A0A9YTC1 A0A0A9YMG1 A0A0A9YMG6
Pubmed
19121390
28756777
26354079
28004739
17994087
24508170
+ More
30249741 18362917 19820115 20798317 21719571 15632085 25348373 21282665 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 29403074 25474469 24495485 25315136 26483478 17510324 25244985 25401762
30249741 18362917 19820115 20798317 21719571 15632085 25348373 21282665 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 29403074 25474469 24495485 25315136 26483478 17510324 25244985 25401762
EMBL
BABH01016761
BABH01016762
ODYU01005832
SOQ47111.1
KZ150025
PZC74811.1
+ More
KQ458761 KPJ05215.1 RSAL01000179 RVE44948.1 GEZM01073729 JAV64906.1 KQ434902 KZC11174.1 CP012526 ALC45955.1 CH916373 EDV94524.1 GALX01005606 JAB62860.1 CH940652 EDW59107.1 KK107019 EZA62767.1 CCAG010000730 JXJN01019898 CH933806 KRG02326.1 EDW16829.1 QOIP01000007 RLU20742.1 KQ971372 EFA10578.2 KQ414581 KOC71040.1 GL451499 EFN79191.1 KK854144 PTY12103.1 OUUW01000005 SPP80691.1 LJIG01022667 KRT79350.1 GL442872 EFN62845.1 GL888184 EGI65635.1 SPP80690.1 SPP80692.1 LBMM01003837 KMQ93067.1 KZ288271 PBC29805.1 KQ976509 KYM82722.1 SPP80689.1 GFTR01008403 JAW08023.1 KQ979039 KYN23357.1 KQ983238 KYQ46211.1 NEVH01017543 PNF23963.1 CM000070 EAL27024.2 CH479186 EDW38867.1 KQ981606 KYN39536.1 GDHF01022597 JAI29717.1 GAKP01012003 JAC46949.1 PNF23968.1 PNF23965.1 GAKP01012007 JAC46945.1 PNF23969.1 ACPB03014352 BT011125 AAR82792.1 PNF23966.1 PNF23967.1 GL762022 EFZ22454.1 PNF23964.1 GBXI01017019 JAC97272.1 GDHF01009708 JAI42606.1 AY051898 AAK93322.1 GFDL01012262 JAV22783.1 GDHF01019450 JAI32864.1 CM000160 EDW98489.2 AE014297 AAF56723.2 AGB96393.1 BT050540 ACJ13247.1 GFDL01012286 JAV22759.1 GFDL01012218 JAV22827.1 JRES01000328 KNC32256.1 CH954182 EDV53398.1 PYGN01001230 PSN36461.1 CH902617 EDV43884.1 GEDC01017218 JAS20080.1 GBBI01002811 JAC15901.1 GAMC01021088 GAMC01021086 JAB85469.1 CH964232 EDW80869.2 KA646154 AFP60783.1 KQ435866 KOX70290.1 JXUM01119268 JXUM01119269 KQ566273 KXJ70260.1 CH477434 EAT40995.1 EAT40994.1 AXCM01002011 DS231964 EDS29455.1 GBHO01010786 JAG32818.1 GBHO01010782 JAG32822.1 GBHO01010784 GBRD01004117 JAG32820.1 JAG61704.1 GBHO01010788 JAG32816.1 GBHO01010783 GBRD01004116 JAG32821.1 JAG61705.1
KQ458761 KPJ05215.1 RSAL01000179 RVE44948.1 GEZM01073729 JAV64906.1 KQ434902 KZC11174.1 CP012526 ALC45955.1 CH916373 EDV94524.1 GALX01005606 JAB62860.1 CH940652 EDW59107.1 KK107019 EZA62767.1 CCAG010000730 JXJN01019898 CH933806 KRG02326.1 EDW16829.1 QOIP01000007 RLU20742.1 KQ971372 EFA10578.2 KQ414581 KOC71040.1 GL451499 EFN79191.1 KK854144 PTY12103.1 OUUW01000005 SPP80691.1 LJIG01022667 KRT79350.1 GL442872 EFN62845.1 GL888184 EGI65635.1 SPP80690.1 SPP80692.1 LBMM01003837 KMQ93067.1 KZ288271 PBC29805.1 KQ976509 KYM82722.1 SPP80689.1 GFTR01008403 JAW08023.1 KQ979039 KYN23357.1 KQ983238 KYQ46211.1 NEVH01017543 PNF23963.1 CM000070 EAL27024.2 CH479186 EDW38867.1 KQ981606 KYN39536.1 GDHF01022597 JAI29717.1 GAKP01012003 JAC46949.1 PNF23968.1 PNF23965.1 GAKP01012007 JAC46945.1 PNF23969.1 ACPB03014352 BT011125 AAR82792.1 PNF23966.1 PNF23967.1 GL762022 EFZ22454.1 PNF23964.1 GBXI01017019 JAC97272.1 GDHF01009708 JAI42606.1 AY051898 AAK93322.1 GFDL01012262 JAV22783.1 GDHF01019450 JAI32864.1 CM000160 EDW98489.2 AE014297 AAF56723.2 AGB96393.1 BT050540 ACJ13247.1 GFDL01012286 JAV22759.1 GFDL01012218 JAV22827.1 JRES01000328 KNC32256.1 CH954182 EDV53398.1 PYGN01001230 PSN36461.1 CH902617 EDV43884.1 GEDC01017218 JAS20080.1 GBBI01002811 JAC15901.1 GAMC01021088 GAMC01021086 JAB85469.1 CH964232 EDW80869.2 KA646154 AFP60783.1 KQ435866 KOX70290.1 JXUM01119268 JXUM01119269 KQ566273 KXJ70260.1 CH477434 EAT40995.1 EAT40994.1 AXCM01002011 DS231964 EDS29455.1 GBHO01010786 JAG32818.1 GBHO01010782 JAG32822.1 GBHO01010784 GBRD01004117 JAG32820.1 JAG61704.1 GBHO01010788 JAG32816.1 GBHO01010783 GBRD01004116 JAG32821.1 JAG61705.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000076502
UP000092553
UP000001070
+ More
UP000008792 UP000053097 UP000092444 UP000092443 UP000092460 UP000092445 UP000009192 UP000078200 UP000091820 UP000005203 UP000279307 UP000007266 UP000053825 UP000008237 UP000192223 UP000268350 UP000000311 UP000007755 UP000036403 UP000242457 UP000078540 UP000078492 UP000075809 UP000235965 UP000001819 UP000008744 UP000078541 UP000095300 UP000015103 UP000192221 UP000002282 UP000000803 UP000079169 UP000037069 UP000008711 UP000245037 UP000007801 UP000007798 UP000075881 UP000095301 UP000053105 UP000069940 UP000249989 UP000008820 UP000075883 UP000075900 UP000076408 UP000075920 UP000002320 UP000075880 UP000075902
UP000008792 UP000053097 UP000092444 UP000092443 UP000092460 UP000092445 UP000009192 UP000078200 UP000091820 UP000005203 UP000279307 UP000007266 UP000053825 UP000008237 UP000192223 UP000268350 UP000000311 UP000007755 UP000036403 UP000242457 UP000078540 UP000078492 UP000075809 UP000235965 UP000001819 UP000008744 UP000078541 UP000095300 UP000015103 UP000192221 UP000002282 UP000000803 UP000079169 UP000037069 UP000008711 UP000245037 UP000007801 UP000007798 UP000075881 UP000095301 UP000053105 UP000069940 UP000249989 UP000008820 UP000075883 UP000075900 UP000076408 UP000075920 UP000002320 UP000075880 UP000075902
Pfam
Interpro
IPR000742
EGF-like_dom
+ More
IPR016201 PSI
IPR011043 Gal_Oxase/kelch_b-propeller
IPR035914 Sperma_CUB_dom_sf
IPR000859 CUB_dom
IPR015915 Kelch-typ_b-propeller
IPR002165 Plexin_repeat
IPR002049 Laminin_EGF
IPR006652 Kelch_1
IPR013032 EGF-like_CS
IPR013111 EGF_extracell
IPR037177 DLC_sf
IPR001372 Dynein_light_chain_typ-1/2
IPR011498 Kelch_2
IPR016201 PSI
IPR011043 Gal_Oxase/kelch_b-propeller
IPR035914 Sperma_CUB_dom_sf
IPR000859 CUB_dom
IPR015915 Kelch-typ_b-propeller
IPR002165 Plexin_repeat
IPR002049 Laminin_EGF
IPR006652 Kelch_1
IPR013032 EGF-like_CS
IPR013111 EGF_extracell
IPR037177 DLC_sf
IPR001372 Dynein_light_chain_typ-1/2
IPR011498 Kelch_2
Gene 3D
CDD
ProteinModelPortal
H9JMR3
A0A2H1W201
A0A2W1BKK8
A0A194QJX6
A0A3S2LVK9
A0A1Y1KZX7
+ More
A0A154PI53 A0A0M4ET48 B4JRY4 V5FYV1 B4M631 A0A026X3D0 A0A1B0FKS7 A0A1A9YEX4 A0A1B0BSW5 A0A1B0A3R6 A0A0Q9X2N6 A0A1A9VGB4 A0A1A9WT77 B4KBA1 A0A088AMS9 A0A3L8DKN8 D6X0Y8 A0A0L7RJI8 E2BYN7 A0A2R7VX70 A0A1W4X0H6 A0A1W4XB92 A0A3B0K6S3 A0A0T6AW74 E2AUP6 F4WJL9 A0A1W4X0E8 A0A3B0JYV9 A0A3B0JK42 A0A0J7KRT9 A0A2A3EDI7 A0A195BEV8 A0A3B0K3X1 A0A224XIR3 A0A195EEX6 A0A151WEJ9 A0A2J7Q5V8 Q29BH4 B4GNY4 A0A195FGN3 A0A0K8USS7 A0A034VWX1 A0A1I8PS18 A0A2J7Q5V6 A0A2J7Q5W5 A0A034VYJ1 A0A2J7Q5W1 T1HMB1 Q6NP06 A0A2J7Q5V1 A0A2J7Q5V2 E9IAJ2 A0A2J7Q5W2 A0A0A1WFU4 A0A0K8VUP9 Q960R8 A0A1Q3F5K1 A0A0K8V1S0 A0A1W4UQ01 B4PRC5 Q9VB20 B6IDV0 A0A1Q3F5F7 A0A1Q3F5J0 A0A3Q0JGC0 A0A0L0CJ26 B3P5Z8 A0A2P8XWR9 B3LXJ0 A0A1B6D320 A0A023F3J4 W8B6R9 B4NAQ5 A0A182JY31 A0A1I8N2W6 T1PBU6 A0A0M8ZSK6 A0A182H8U6 Q172L4 Q172L3 A0A182LX91 A0A182R891 A0A182YLM0 A0A182VS49 B0WJT8 A0A0A9YNY9 A0A182JLU8 A0A0A9YIC6 A0A182TLW8 A0A0A9YTC1 A0A0A9YMG1 A0A0A9YMG6
A0A154PI53 A0A0M4ET48 B4JRY4 V5FYV1 B4M631 A0A026X3D0 A0A1B0FKS7 A0A1A9YEX4 A0A1B0BSW5 A0A1B0A3R6 A0A0Q9X2N6 A0A1A9VGB4 A0A1A9WT77 B4KBA1 A0A088AMS9 A0A3L8DKN8 D6X0Y8 A0A0L7RJI8 E2BYN7 A0A2R7VX70 A0A1W4X0H6 A0A1W4XB92 A0A3B0K6S3 A0A0T6AW74 E2AUP6 F4WJL9 A0A1W4X0E8 A0A3B0JYV9 A0A3B0JK42 A0A0J7KRT9 A0A2A3EDI7 A0A195BEV8 A0A3B0K3X1 A0A224XIR3 A0A195EEX6 A0A151WEJ9 A0A2J7Q5V8 Q29BH4 B4GNY4 A0A195FGN3 A0A0K8USS7 A0A034VWX1 A0A1I8PS18 A0A2J7Q5V6 A0A2J7Q5W5 A0A034VYJ1 A0A2J7Q5W1 T1HMB1 Q6NP06 A0A2J7Q5V1 A0A2J7Q5V2 E9IAJ2 A0A2J7Q5W2 A0A0A1WFU4 A0A0K8VUP9 Q960R8 A0A1Q3F5K1 A0A0K8V1S0 A0A1W4UQ01 B4PRC5 Q9VB20 B6IDV0 A0A1Q3F5F7 A0A1Q3F5J0 A0A3Q0JGC0 A0A0L0CJ26 B3P5Z8 A0A2P8XWR9 B3LXJ0 A0A1B6D320 A0A023F3J4 W8B6R9 B4NAQ5 A0A182JY31 A0A1I8N2W6 T1PBU6 A0A0M8ZSK6 A0A182H8U6 Q172L4 Q172L3 A0A182LX91 A0A182R891 A0A182YLM0 A0A182VS49 B0WJT8 A0A0A9YNY9 A0A182JLU8 A0A0A9YIC6 A0A182TLW8 A0A0A9YTC1 A0A0A9YMG1 A0A0A9YMG6
Ontologies
GO
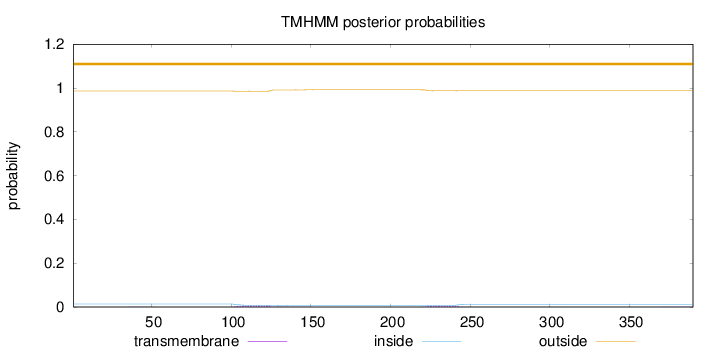
Topology
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27604
Exp number, first 60 AAs:
0.00288
Total prob of N-in:
0.01378
outside
1 - 390
Population Genetic Test Statistics
Pi
196.311144
Theta
152.995443
Tajima's D
1.495403
CLR
0
CSRT
0.789060546972651
Interpretation
Uncertain
