Gene
KWMTBOMO15549 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010738
Annotation
PREDICTED:_retinol_dehydrogenase_14-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.921
Sequence
CDS
ATGATTGTCGCGGAATTCTGGAAAGTGAGTTACCTTTGGTGGCTCCTCCCGGCTCTCATCATAGCTGGCCTGATCGCCATACTAGGTCTCATTGGTGTACTCTTCGCCTTACATCTGTACGTTAAGTGGACTTGTGGTATTTACAACGGGAAAACCAAAATGGACGGGAAGACGGTAATAGTTACAGGATGCACTAGCGGAATAGGAAAGGAAACCGCCAGGGAATTGGCTCAGAGAGGCGCTCGAGTCATAATGGCCTGCAGGAATATGGACGCCGCTGAGAAAGTCAAAGCTGAAATCATCGAAACAACTAGAAACACCAAAATAGAAGTCAAGAAACTCGATCTCAGTTCATTCGCGTCTATCAGAGAATTCGCTAAAGACATCAACAAAACAGAAGCCCGTCTGGATGTCCTTGTGCACAACGCAGGATACGCCGAAACATTTAAAAAGAACACAACAGTTGATAACCTCGAAATGACTATGGCCACAAACCATTACGGTCCCTTTTTACTCACTCATCTGCTGATAGATTTATTGAAGAAAACCGCGCCCTCGAGAATCGTTATAGTAGCATCCTCTTTATACAGACTTGCTTCGTTTGACTTAAACAAACCGAACCCTGTGGATTCGATTCCAGGGTATCTGTACTACGCTTCCAAAGAAGCGAATATTCTGTTCACAAAAGAATTGGCCCGGAGATTGGAAGGTACTGGGGTTACAGCTAACTGCCTGCACCCTGGACTAATCGATACGGGCATCTGGAGATCAGTCCCGGCACCATTAAGTTGGGGCTTAAACTTGATTAACAAATGCTTCTTTAAGACTCCAGAGCAGGGTTGCCAGACTACAATAATGTTAGCAGTTGATGAAAGTTTAGAGAAAGTGAACGGAAAATACTTTTCTGACTGTCAGGAGAGTTCGGTGTCTCAATCAGCTAGTGACATGGCAAAAGCTAGGAAATTATGGGAGATATCTGAGAAAATAGTTAATCTTGAGGAGACTGACCCAAGAATTTTGTAA
Protein
MIVAEFWKVSYLWWLLPALIIAGLIAILGLIGVLFALHLYVKWTCGIYNGKTKMDGKTVIVTGCTSGIGKETARELAQRGARVIMACRNMDAAEKVKAEIIETTRNTKIEVKKLDLSSFASIREFAKDINKTEARLDVLVHNAGYAETFKKNTTVDNLEMTMATNHYGPFLLTHLLIDLLKKTAPSRIVIVASSLYRLASFDLNKPNPVDSIPGYLYYASKEANILFTKELARRLEGTGVTANCLHPGLIDTGIWRSVPAPLSWGLNLINKCFFKTPEQGCQTTIMLAVDESLEKVNGKYFSDCQESSVSQSASDMAKARKLWEISEKIVNLEETDPRIL
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JMI5
A0A2W1BQ73
A0A1Q3G5I9
A0A2A4J204
A0A0L7LA56
A0A2H1VPB2
+ More
S4PC03 A0A212EHQ0 I4DJW3 A0A3S2LVK4 A0A194PLT0 A0A0N0PDQ0 A0A1B6LG81 K7IP74 A0A1B6FRX7 A0A1B6IV52 A0A232FAM6 A0A151IL06 E2AM66 A0A195FH86 A0A151IVL0 A0A026WH33 E2BNF8 A0A195BEN0 A0A158NKK8 A0A151WEM9 A0A1B6DPI9 A0A0J7L004 A0A1W4XHF8 F4X379 A0A2M4AP46 A0A182U7J4 A0A182PZY6 A0A182VIF5 A0A182LH67 A0A182HY29 A0A0L7RHT2 Q17GA4 A0A1S4H6E5 A0A310SPI3 A0A1Y1KEI4 A0A0T6ATX4 A0A182IYN9 J3JZD6 A0A0M9A5H7 B0XLA9 A0A084VI03 A0A0K8T4Q4 A0A088AQ58 A0A0A9YH42 A0A2H8TYL3 A0A154P7Q6 A0A182P388 A0A2S2QMQ3 J9K599 A0A182WXF2 A0A182MFP7 A0A2P8XZE3 A0A182N3A3 A0A069DST5 B4M3Z8 A0A2J7QDU2 A0A182RVG0 A0A182W1P5 A0A1J1J013 A0A034W5L3 A0A0K8URJ7 A0A224XFL4 B4K931 A0A1Q3FPZ3 A0A0Q9WC99 A0A3B0K5R4 R4FLL5 A0A1J1HPR2 B4JF21 A0A0A1XDP5 D6X2D0 B4G5D3 A0A0Q9WYL5 A0A034W3X1 A0A0K8VZD3 A0A0M4EPM4 A0A023F7P3 A0A3B0K400 A0A182KH00 A0A1B0CS46 A0A146M601 A0A182FBX6 W8BZ25 A0A0L0C6N2 A0A067RFS8 Q9VE80 T1PIN4 B3NZ64 A0A1J1J3F9 A0A1I8P8Z8 A0A0A1X7Y5 B4QUF1 B4PLB8 A0A336MN15
S4PC03 A0A212EHQ0 I4DJW3 A0A3S2LVK4 A0A194PLT0 A0A0N0PDQ0 A0A1B6LG81 K7IP74 A0A1B6FRX7 A0A1B6IV52 A0A232FAM6 A0A151IL06 E2AM66 A0A195FH86 A0A151IVL0 A0A026WH33 E2BNF8 A0A195BEN0 A0A158NKK8 A0A151WEM9 A0A1B6DPI9 A0A0J7L004 A0A1W4XHF8 F4X379 A0A2M4AP46 A0A182U7J4 A0A182PZY6 A0A182VIF5 A0A182LH67 A0A182HY29 A0A0L7RHT2 Q17GA4 A0A1S4H6E5 A0A310SPI3 A0A1Y1KEI4 A0A0T6ATX4 A0A182IYN9 J3JZD6 A0A0M9A5H7 B0XLA9 A0A084VI03 A0A0K8T4Q4 A0A088AQ58 A0A0A9YH42 A0A2H8TYL3 A0A154P7Q6 A0A182P388 A0A2S2QMQ3 J9K599 A0A182WXF2 A0A182MFP7 A0A2P8XZE3 A0A182N3A3 A0A069DST5 B4M3Z8 A0A2J7QDU2 A0A182RVG0 A0A182W1P5 A0A1J1J013 A0A034W5L3 A0A0K8URJ7 A0A224XFL4 B4K931 A0A1Q3FPZ3 A0A0Q9WC99 A0A3B0K5R4 R4FLL5 A0A1J1HPR2 B4JF21 A0A0A1XDP5 D6X2D0 B4G5D3 A0A0Q9WYL5 A0A034W3X1 A0A0K8VZD3 A0A0M4EPM4 A0A023F7P3 A0A3B0K400 A0A182KH00 A0A1B0CS46 A0A146M601 A0A182FBX6 W8BZ25 A0A0L0C6N2 A0A067RFS8 Q9VE80 T1PIN4 B3NZ64 A0A1J1J3F9 A0A1I8P8Z8 A0A0A1X7Y5 B4QUF1 B4PLB8 A0A336MN15
Pubmed
19121390
28756777
26227816
23622113
22118469
22651552
+ More
26354079 20075255 28648823 20798317 24508170 30249741 21347285 21719571 20966253 17510324 12364791 28004739 22516182 23537049 24438588 26823975 25401762 29403074 26334808 17994087 25348373 25830018 18362917 19820115 25474469 24495485 26108605 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304
26354079 20075255 28648823 20798317 24508170 30249741 21347285 21719571 20966253 17510324 12364791 28004739 22516182 23537049 24438588 26823975 25401762 29403074 26334808 17994087 25348373 25830018 18362917 19820115 25474469 24495485 26108605 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304
EMBL
BABH01016712
KZ149936
PZC77138.1
GEYN01000193
JAV44796.1
NWSH01003845
+ More
PCG65766.1 JTDY01002030 KOB72275.1 ODYU01003647 SOQ42663.1 GAIX01004371 JAA88189.1 AGBW02014829 OWR41009.1 AK401581 BAM18203.1 RSAL01000165 RVE45291.1 KQ459601 KPI93694.1 KQ460129 KPJ17547.1 GEBQ01017270 JAT22707.1 GECZ01025002 GECZ01017006 JAS44767.1 JAS52763.1 GECU01016890 JAS90816.1 NNAY01000591 OXU27509.1 KQ977151 KYN05278.1 GL440715 EFN65472.1 KQ981606 KYN39592.1 KQ980895 KYN11766.1 KK107250 QOIP01000002 EZA54384.1 RLU25994.1 GL449414 EFN82737.1 KQ976509 KYM82669.1 ADTU01000291 KQ983238 KYQ46262.1 GEDC01009721 JAS27577.1 LBMM01001639 KMQ95961.1 GL888609 EGI59060.1 GGFK01009206 MBW42527.1 AXCN02001120 APCN01004485 KQ414590 KOC70369.1 CH477264 EAT45601.1 EAT45602.1 AAAB01008816 KQ760478 OAD60211.1 GEZM01085981 GEZM01085980 GEZM01085979 GEZM01085978 GEZM01085977 GEZM01085976 JAV59859.1 LJIG01022860 KRT78347.1 APGK01059021 BT128617 KB741292 KB631582 AEE63574.1 ENN70380.1 ERL84491.1 KQ435739 KOX76876.1 DS234386 EDS33797.1 ATLV01013254 KE524847 KFB37597.1 GBRD01005410 GDHC01002213 JAG60411.1 JAQ16416.1 GBHO01012663 GBHO01002098 JAG30941.1 JAG41506.1 GFXV01007598 MBW19403.1 KQ434839 KZC07955.1 GGMS01009768 MBY78971.1 ABLF02036064 AXCM01007670 PYGN01001126 PSN37382.1 GBGD01002103 JAC86786.1 CH940652 EDW59359.1 NEVH01015814 PNF26749.1 CVRI01000064 CRL05140.1 GAKP01009537 JAC49415.1 GDHF01023108 JAI29206.1 GFTR01005159 JAW11267.1 CH933806 EDW14444.1 GFDL01005380 JAV29665.1 KRF78815.1 OUUW01000013 SPP88012.1 GAHY01001333 JAA76177.1 CVRI01000006 CRK88201.1 CH916369 EDV93302.1 GBXI01016837 GBXI01005231 JAC97454.1 JAD09061.1 KQ971371 EFA09879.2 CH479179 EDW24799.1 KRG01019.1 GAKP01009538 JAC49414.1 GDHF01008354 JAI43960.1 CP012526 ALC46097.1 GBBI01001321 JAC17391.1 SPP88013.1 AJWK01025642 GDHC01004443 JAQ14186.1 GAMC01004622 JAC01934.1 JRES01000835 KNC27925.1 KK852714 KDR17887.1 AE014297 AAF55546.2 AHN57389.1 KA648636 AFP63265.1 CH954181 EDV48606.1 CRL05417.1 GBXI01006878 JAD07414.1 CM000364 EDX12451.1 CM000160 EDW95900.1 UFQT01001800 SSX31834.1
PCG65766.1 JTDY01002030 KOB72275.1 ODYU01003647 SOQ42663.1 GAIX01004371 JAA88189.1 AGBW02014829 OWR41009.1 AK401581 BAM18203.1 RSAL01000165 RVE45291.1 KQ459601 KPI93694.1 KQ460129 KPJ17547.1 GEBQ01017270 JAT22707.1 GECZ01025002 GECZ01017006 JAS44767.1 JAS52763.1 GECU01016890 JAS90816.1 NNAY01000591 OXU27509.1 KQ977151 KYN05278.1 GL440715 EFN65472.1 KQ981606 KYN39592.1 KQ980895 KYN11766.1 KK107250 QOIP01000002 EZA54384.1 RLU25994.1 GL449414 EFN82737.1 KQ976509 KYM82669.1 ADTU01000291 KQ983238 KYQ46262.1 GEDC01009721 JAS27577.1 LBMM01001639 KMQ95961.1 GL888609 EGI59060.1 GGFK01009206 MBW42527.1 AXCN02001120 APCN01004485 KQ414590 KOC70369.1 CH477264 EAT45601.1 EAT45602.1 AAAB01008816 KQ760478 OAD60211.1 GEZM01085981 GEZM01085980 GEZM01085979 GEZM01085978 GEZM01085977 GEZM01085976 JAV59859.1 LJIG01022860 KRT78347.1 APGK01059021 BT128617 KB741292 KB631582 AEE63574.1 ENN70380.1 ERL84491.1 KQ435739 KOX76876.1 DS234386 EDS33797.1 ATLV01013254 KE524847 KFB37597.1 GBRD01005410 GDHC01002213 JAG60411.1 JAQ16416.1 GBHO01012663 GBHO01002098 JAG30941.1 JAG41506.1 GFXV01007598 MBW19403.1 KQ434839 KZC07955.1 GGMS01009768 MBY78971.1 ABLF02036064 AXCM01007670 PYGN01001126 PSN37382.1 GBGD01002103 JAC86786.1 CH940652 EDW59359.1 NEVH01015814 PNF26749.1 CVRI01000064 CRL05140.1 GAKP01009537 JAC49415.1 GDHF01023108 JAI29206.1 GFTR01005159 JAW11267.1 CH933806 EDW14444.1 GFDL01005380 JAV29665.1 KRF78815.1 OUUW01000013 SPP88012.1 GAHY01001333 JAA76177.1 CVRI01000006 CRK88201.1 CH916369 EDV93302.1 GBXI01016837 GBXI01005231 JAC97454.1 JAD09061.1 KQ971371 EFA09879.2 CH479179 EDW24799.1 KRG01019.1 GAKP01009538 JAC49414.1 GDHF01008354 JAI43960.1 CP012526 ALC46097.1 GBBI01001321 JAC17391.1 SPP88013.1 AJWK01025642 GDHC01004443 JAQ14186.1 GAMC01004622 JAC01934.1 JRES01000835 KNC27925.1 KK852714 KDR17887.1 AE014297 AAF55546.2 AHN57389.1 KA648636 AFP63265.1 CH954181 EDV48606.1 CRL05417.1 GBXI01006878 JAD07414.1 CM000364 EDX12451.1 CM000160 EDW95900.1 UFQT01001800 SSX31834.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000283053
UP000053268
+ More
UP000053240 UP000002358 UP000215335 UP000078542 UP000000311 UP000078541 UP000078492 UP000053097 UP000279307 UP000008237 UP000078540 UP000005205 UP000075809 UP000036403 UP000192223 UP000007755 UP000075902 UP000075886 UP000075903 UP000075882 UP000075840 UP000053825 UP000008820 UP000075880 UP000019118 UP000030742 UP000053105 UP000002320 UP000030765 UP000005203 UP000076502 UP000075885 UP000007819 UP000076407 UP000075883 UP000245037 UP000075884 UP000008792 UP000235965 UP000075900 UP000075920 UP000183832 UP000009192 UP000268350 UP000001070 UP000007266 UP000008744 UP000092553 UP000075881 UP000092461 UP000069272 UP000037069 UP000027135 UP000000803 UP000095301 UP000008711 UP000095300 UP000000304 UP000002282
UP000053240 UP000002358 UP000215335 UP000078542 UP000000311 UP000078541 UP000078492 UP000053097 UP000279307 UP000008237 UP000078540 UP000005205 UP000075809 UP000036403 UP000192223 UP000007755 UP000075902 UP000075886 UP000075903 UP000075882 UP000075840 UP000053825 UP000008820 UP000075880 UP000019118 UP000030742 UP000053105 UP000002320 UP000030765 UP000005203 UP000076502 UP000075885 UP000007819 UP000076407 UP000075883 UP000245037 UP000075884 UP000008792 UP000235965 UP000075900 UP000075920 UP000183832 UP000009192 UP000268350 UP000001070 UP000007266 UP000008744 UP000092553 UP000075881 UP000092461 UP000069272 UP000037069 UP000027135 UP000000803 UP000095301 UP000008711 UP000095300 UP000000304 UP000002282
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JMI5
A0A2W1BQ73
A0A1Q3G5I9
A0A2A4J204
A0A0L7LA56
A0A2H1VPB2
+ More
S4PC03 A0A212EHQ0 I4DJW3 A0A3S2LVK4 A0A194PLT0 A0A0N0PDQ0 A0A1B6LG81 K7IP74 A0A1B6FRX7 A0A1B6IV52 A0A232FAM6 A0A151IL06 E2AM66 A0A195FH86 A0A151IVL0 A0A026WH33 E2BNF8 A0A195BEN0 A0A158NKK8 A0A151WEM9 A0A1B6DPI9 A0A0J7L004 A0A1W4XHF8 F4X379 A0A2M4AP46 A0A182U7J4 A0A182PZY6 A0A182VIF5 A0A182LH67 A0A182HY29 A0A0L7RHT2 Q17GA4 A0A1S4H6E5 A0A310SPI3 A0A1Y1KEI4 A0A0T6ATX4 A0A182IYN9 J3JZD6 A0A0M9A5H7 B0XLA9 A0A084VI03 A0A0K8T4Q4 A0A088AQ58 A0A0A9YH42 A0A2H8TYL3 A0A154P7Q6 A0A182P388 A0A2S2QMQ3 J9K599 A0A182WXF2 A0A182MFP7 A0A2P8XZE3 A0A182N3A3 A0A069DST5 B4M3Z8 A0A2J7QDU2 A0A182RVG0 A0A182W1P5 A0A1J1J013 A0A034W5L3 A0A0K8URJ7 A0A224XFL4 B4K931 A0A1Q3FPZ3 A0A0Q9WC99 A0A3B0K5R4 R4FLL5 A0A1J1HPR2 B4JF21 A0A0A1XDP5 D6X2D0 B4G5D3 A0A0Q9WYL5 A0A034W3X1 A0A0K8VZD3 A0A0M4EPM4 A0A023F7P3 A0A3B0K400 A0A182KH00 A0A1B0CS46 A0A146M601 A0A182FBX6 W8BZ25 A0A0L0C6N2 A0A067RFS8 Q9VE80 T1PIN4 B3NZ64 A0A1J1J3F9 A0A1I8P8Z8 A0A0A1X7Y5 B4QUF1 B4PLB8 A0A336MN15
S4PC03 A0A212EHQ0 I4DJW3 A0A3S2LVK4 A0A194PLT0 A0A0N0PDQ0 A0A1B6LG81 K7IP74 A0A1B6FRX7 A0A1B6IV52 A0A232FAM6 A0A151IL06 E2AM66 A0A195FH86 A0A151IVL0 A0A026WH33 E2BNF8 A0A195BEN0 A0A158NKK8 A0A151WEM9 A0A1B6DPI9 A0A0J7L004 A0A1W4XHF8 F4X379 A0A2M4AP46 A0A182U7J4 A0A182PZY6 A0A182VIF5 A0A182LH67 A0A182HY29 A0A0L7RHT2 Q17GA4 A0A1S4H6E5 A0A310SPI3 A0A1Y1KEI4 A0A0T6ATX4 A0A182IYN9 J3JZD6 A0A0M9A5H7 B0XLA9 A0A084VI03 A0A0K8T4Q4 A0A088AQ58 A0A0A9YH42 A0A2H8TYL3 A0A154P7Q6 A0A182P388 A0A2S2QMQ3 J9K599 A0A182WXF2 A0A182MFP7 A0A2P8XZE3 A0A182N3A3 A0A069DST5 B4M3Z8 A0A2J7QDU2 A0A182RVG0 A0A182W1P5 A0A1J1J013 A0A034W5L3 A0A0K8URJ7 A0A224XFL4 B4K931 A0A1Q3FPZ3 A0A0Q9WC99 A0A3B0K5R4 R4FLL5 A0A1J1HPR2 B4JF21 A0A0A1XDP5 D6X2D0 B4G5D3 A0A0Q9WYL5 A0A034W3X1 A0A0K8VZD3 A0A0M4EPM4 A0A023F7P3 A0A3B0K400 A0A182KH00 A0A1B0CS46 A0A146M601 A0A182FBX6 W8BZ25 A0A0L0C6N2 A0A067RFS8 Q9VE80 T1PIN4 B3NZ64 A0A1J1J3F9 A0A1I8P8Z8 A0A0A1X7Y5 B4QUF1 B4PLB8 A0A336MN15
PDB
3RD5
E-value=3.21819e-23,
Score=267
Ontologies
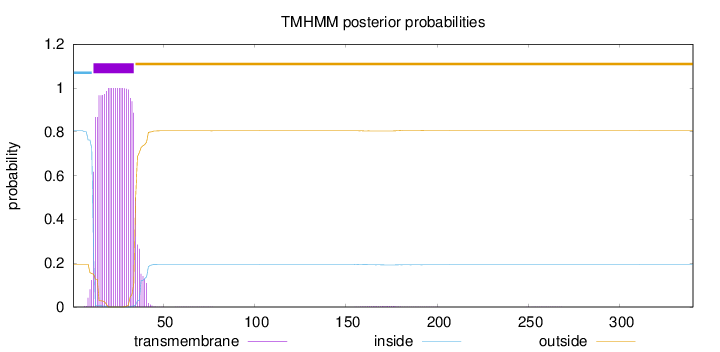
Topology
Length:
340
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.94972
Exp number, first 60 AAs:
23.85421
Total prob of N-in:
0.80450
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 340
Population Genetic Test Statistics
Pi
127.691814
Theta
122.422284
Tajima's D
-0.333821
CLR
0.545922
CSRT
0.285785710714464
Interpretation
Uncertain
