Pre Gene Modal
BGIBMGA010804
Annotation
PREDICTED:_DNA-directed_RNA_polymerase_I_subunit_RPA2_[Papilio_polytes]
Full name
DNA-directed RNA polymerase subunit beta
Location in the cell
Cytoplasmic Reliability : 1.177 Mitochondrial Reliability : 1.416
Sequence
CDS
ATGAATCCAAAAAAACTAGCCCAAAACCCATCATTGGCGTACACAAGCCACCCAGATTATAGAAAACCGCCAAAAATAGCAAATCCGTACCTTCAAAGTCTTGGAGCGCCTCACATAGATTCATTCAATTATATGCTGGACGATGGCCTGAAATTTGCTATAGCTGATCTATTGCCATCGGAATTTGAGTTACCTAGTGGTGAAAAGGTCAAAGTGACAATCGATGAAGCAGCATTCGCTAAACCCAATGTTCCCATGGATACAGTAGGGGTTAAATCTCAAGTGGTCCTCCCAACAGAATGCCGGCAGAGAGCTGCTACATACAAGGGAGAGTTAAAGATCAGAGTCACTCTGTGCATTGACGGAAGGTCTGTCACCATTGAGAGGTCCTTAGGTTATTTACCCATTATGATTAAGTCTAAAATGTGTCATTTGGCTGATTTATCACCTGAAGAACTGATTGAGAAAAATGAACATGCTGATGAATGGGGTGGATATTTTGTAATAAAGGGCCATGAGCGTCTAGCCCGTATGTTGTTGGTTACACGAAGGAACTACCCTGTGGCCATCAAGCGGTCAGGATGGAAGATGCGAGGAAATCTCTTCTCAGACTATGGAGTATTAGTACGTTGCGTCACCTCCGATCAGACCAGCACGAACAATGTTCTGCATTTCCTAACGAATGGCACGTGCAAGCTGATGTTCTCGTACCGCAAGATGATGTACTACGCTCCCCTACTCCTCATAATGAAGTGCCTGGTCCCGTGGCCGGATCACTACATATACACACTGCTGTTGCAGCATAATAAGAATGATTTGTACTACGTCAATTGCATCCAGAATATGCTCCGAGAGCTCCACGAGGAAGGTCTGCACACATCCGAAGAGTGTCGCCTGTACTTAGGCAAGATGTTCCGCTCCAAGCTGTACGAGCTGCCGCCCTGGGCTACCGAGATGGAGGCGGCCCAGTTCCTCCTGGACCGATGCGTTATGATCCACCTAAAGAACAACACGGACAAGTTCCACGCTTTAGTCTTCATGGTCCAGAAGCTCTACGATCTGGTTCAGAATAAGTGTAAGGTGGAAGGAGCGGATGCTGTAATGATGCAGGAGGTGATGGTGGGAGGCCACCTCTACTTGCAGGTGTTGAAGGAAAGACTCCAGAATATTCTCAATAACGTCAGGATACAGATCCTTAAACGCGCCAAGACAACACATCGACTAGTCATCGGTCATAAAGAGCTTCAACAAATCCTCCGCGCGTGCGGGAGTCTAGAGCAGAAGATGGAGACGTTCTTGGCCACGGGGAATGCGCCCTCCAGCAACGTCAACCTCACGCAGTACAAGGGACTCACCATCGTGGCCGAGAACATCAACAGGATGCGATATATGTCGCATTTCAAGTCGATTCACCGCGGCTCGTTCTTCATGGAGATGCGCACGACGGAGGCCCGGCAGCTGCTGCCGGACGCGTGGGGCTTCGTGTGTCCCGTGCACACGCCCGACGGCGCGCCCTGCGGACTGCTCAACCACCTCACCATGTCCGCGCAGATCACACAAATTCCCGATCAAAAATTAGTGGATAATCTTCCTTTAGTTTTACAAAACTGCGGTATGGAGCCCATAAGCAGCGTGCTCTGCGACCGCGACACGTACAAGTACCCGGTGTTCGTGGACGGGCGGCTGGTGGGCTACATCGCGGAGAGCGGCGCCGAGAAGTCGGCCGCCTACCTCCGCACCCTCAAGATAAAGGGCGAAGAGGTTCCGATCACTACCGAGATTGTGGTCATCCCTAAAAAACAGATCTGCGCCCAATACCCTGGCATATTCCTGATGACGACGGAGGCCCGCATGATGCGGCCGGTCATCAACCTGGCCACCTCGCAGTTAGAGCTGATCGGTACAATGGAGCAGCTGTACCTGGATATCGCCATAACGCCGAACGAGGTTCACAAAGGTCAGACGACGCACATGGAGCTTTCGCAGTCAGCGTTCCTGAGCAACTTGGCGCAGCTGGTGCCGATGCCGGACTGTAATCAGTCTCCAAGGAACATGTACCAGTGCCAGATGGGCAAGCAGACGATGGGCACCCCGGTGCACACGTGGGGCGGCACCGGCCCCGGCAAGCTGTACCGGCTGCAGACTCCCGCCGCGCCGCTGTTCCGCCCCGCGCACCACGACCGCCTCGCCCTCGACGACTACCCCGCCGGGACCAACGCCATCGTCGCCGTCATATCCTACACGGTCAGTACCCCTAACCTGACCTTGAACCCTTAG
Protein
MNPKKLAQNPSLAYTSHPDYRKPPKIANPYLQSLGAPHIDSFNYMLDDGLKFAIADLLPSEFELPSGEKVKVTIDEAAFAKPNVPMDTVGVKSQVVLPTECRQRAATYKGELKIRVTLCIDGRSVTIERSLGYLPIMIKSKMCHLADLSPEELIEKNEHADEWGGYFVIKGHERLARMLLVTRRNYPVAIKRSGWKMRGNLFSDYGVLVRCVTSDQTSTNNVLHFLTNGTCKLMFSYRKMMYYAPLLLIMKCLVPWPDHYIYTLLLQHNKNDLYYVNCIQNMLRELHEEGLHTSEECRLYLGKMFRSKLYELPPWATEMEAAQFLLDRCVMIHLKNNTDKFHALVFMVQKLYDLVQNKCKVEGADAVMMQEVMVGGHLYLQVLKERLQNILNNVRIQILKRAKTTHRLVIGHKELQQILRACGSLEQKMETFLATGNAPSSNVNLTQYKGLTIVAENINRMRYMSHFKSIHRGSFFMEMRTTEARQLLPDAWGFVCPVHTPDGAPCGLLNHLTMSAQITQIPDQKLVDNLPLVLQNCGMEPISSVLCDRDTYKYPVFVDGRLVGYIAESGAEKSAAYLRTLKIKGEEVPITTEIVVIPKKQICAQYPGIFLMTTEARMMRPVINLATSQLELIGTMEQLYLDIAITPNEVHKGQTTHMELSQSAFLSNLAQLVPMPDCNQSPRNMYQCQMGKQTMGTPVHTWGGTGPGKLYRLQTPAAPLFRPAHHDRLALDDYPAGTNAIVAVISYTVSTPNLTLNP
Summary
Description
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates.
Catalytic Activity
a ribonucleoside 5'-triphosphate + RNA(n) = diphosphate + RNA(n+1)
Similarity
Belongs to the RNA polymerase beta chain family.
Uniprot
H9JMQ1
A0A0N1ICC8
A0A212FAC3
A0A3S2NX40
A0A2W1BZB1
A0A212F425
+ More
A0A1W4XHB8 A0A154NVW8 A0A088A2A1 A0A232EXH0 A0A2A3EBT9 A0A1Y1KKJ0 K7IWJ3 N6TC52 E2B687 A0A0J7NJI8 A0A1B6M535 F4WB73 A0A1B6G6E8 A0A023F422 A0A2J7Q9B5 E9IAZ0 A0A158P3I7 A0A1B6K312 A0A1B6CSM5 A0A151JXF7 A0A1B6CFG7 A0A0P4VRX0 U4U813 A0A0V0G7L3 A0A151J8Q7 A0A151HZI2 A0A026WUS5 A0A224XGY9 A0A151IL56 J9E9J9 A0A182GW05 A0A182GJ47 A0A224XFY6 A0A0L0CDA1 A0A1I8QCQ3 D6WVZ3 E0V8Y6 A0A151XDM1 A0A1Q3EXN9 A0A1I8M669 A0A067QQ90 A0A0P5QZ94 A0A0N8E7P0 A0A0P5L5Y1 A0A0P6GD02 J9K6G2 W8B904 A0A146LR30 A0A146LJQ4 A0A0A9VR72 B0W5T2 A0A1J1HL88 A0A0P6IJK5 A0A0P4ZUJ3 A0A2H8TU91 A0A0A1XSL8 A0A0K8UKU8 A0A034WES4 A0A1A9V0X0 A0A1B0FD37 A0A1A9YSF8 A0A1A9ZQR0 A0A2S2QW78 A0A1A9WUR2 A0A2M4CVF7 A0A2M4BD15 A0A2M4CU57 A0A2M4BDL2 A0A2M4BCX8 W5J6H6 A0A2M4AHX0 A0A2M4AIY3 A0A0L7R3C5 A0A182L5Q9 Q7QG74 A0A182F4E3 A0A3Q0ITX4 A0A182X7Q7 A0A182IE12 A0A182NE81 A0A182RGH5 E9HT05 A0A182VG37 A0A084VPA9 A0A182TZ99 A0A182MM49 A0A0K2TFZ6 A0A182P955 B4KFJ3 B3MUI2 B4N0U3 A0A182Q1G0 A0A182JYE9 A0A182VV21
A0A1W4XHB8 A0A154NVW8 A0A088A2A1 A0A232EXH0 A0A2A3EBT9 A0A1Y1KKJ0 K7IWJ3 N6TC52 E2B687 A0A0J7NJI8 A0A1B6M535 F4WB73 A0A1B6G6E8 A0A023F422 A0A2J7Q9B5 E9IAZ0 A0A158P3I7 A0A1B6K312 A0A1B6CSM5 A0A151JXF7 A0A1B6CFG7 A0A0P4VRX0 U4U813 A0A0V0G7L3 A0A151J8Q7 A0A151HZI2 A0A026WUS5 A0A224XGY9 A0A151IL56 J9E9J9 A0A182GW05 A0A182GJ47 A0A224XFY6 A0A0L0CDA1 A0A1I8QCQ3 D6WVZ3 E0V8Y6 A0A151XDM1 A0A1Q3EXN9 A0A1I8M669 A0A067QQ90 A0A0P5QZ94 A0A0N8E7P0 A0A0P5L5Y1 A0A0P6GD02 J9K6G2 W8B904 A0A146LR30 A0A146LJQ4 A0A0A9VR72 B0W5T2 A0A1J1HL88 A0A0P6IJK5 A0A0P4ZUJ3 A0A2H8TU91 A0A0A1XSL8 A0A0K8UKU8 A0A034WES4 A0A1A9V0X0 A0A1B0FD37 A0A1A9YSF8 A0A1A9ZQR0 A0A2S2QW78 A0A1A9WUR2 A0A2M4CVF7 A0A2M4BD15 A0A2M4CU57 A0A2M4BDL2 A0A2M4BCX8 W5J6H6 A0A2M4AHX0 A0A2M4AIY3 A0A0L7R3C5 A0A182L5Q9 Q7QG74 A0A182F4E3 A0A3Q0ITX4 A0A182X7Q7 A0A182IE12 A0A182NE81 A0A182RGH5 E9HT05 A0A182VG37 A0A084VPA9 A0A182TZ99 A0A182MM49 A0A0K2TFZ6 A0A182P955 B4KFJ3 B3MUI2 B4N0U3 A0A182Q1G0 A0A182JYE9 A0A182VV21
EC Number
2.7.7.6
Pubmed
19121390
26354079
22118469
28756777
28648823
28004739
+ More
20075255 23537049 20798317 21719571 25474469 21282665 21347285 27129103 24508170 30249741 17510324 26483478 26108605 18362917 19820115 20566863 25315136 24845553 24495485 26823975 25401762 25830018 25348373 20920257 23761445 20966253 12364791 21292972 24438588 17994087
20075255 23537049 20798317 21719571 25474469 21282665 21347285 27129103 24508170 30249741 17510324 26483478 26108605 18362917 19820115 20566863 25315136 24845553 24495485 26823975 25401762 25830018 25348373 20920257 23761445 20966253 12364791 21292972 24438588 17994087
EMBL
BABH01016707
BABH01016708
BABH01016709
BABH01016710
KQ459910
KPJ19329.1
+ More
AGBW02009499 OWR50682.1 RSAL01000118 RVE46764.1 KZ149936 PZC77143.1 AGBW02010450 OWR48487.1 KQ434769 KZC03787.1 NNAY01001730 OXU23104.1 KZ288311 PBC28511.1 GEZM01085172 GEZM01085171 GEZM01085170 JAV60155.1 APGK01035628 APGK01035629 APGK01035630 KB740928 ENN77884.1 GL445930 EFN88780.1 LBMM01004251 KMQ92660.1 GEBQ01008946 JAT31031.1 GL888058 EGI68546.1 GECZ01011759 JAS58010.1 GBBI01002988 JAC15724.1 NEVH01016943 PNF25173.1 GL762091 EFZ22265.1 ADTU01008085 ADTU01008086 ADTU01008087 ADTU01008088 GECU01001857 JAT05850.1 GEDC01021045 JAS16253.1 KQ981604 KYN39719.1 GEDC01025130 JAS12168.1 GDKW01000777 JAI55818.1 KB632064 ERL88468.1 GECL01002690 JAP03434.1 KQ979488 KYN21228.1 KQ976710 KYM77038.1 KK107088 QOIP01000008 EZA59800.1 RLU19525.1 GFTR01008584 JAW07842.1 KQ977133 KYN05487.1 CH477305 EJY57536.1 JXUM01092462 KQ563989 KXJ72968.1 JXUM01067297 KQ562443 KXJ75905.1 GFTR01008924 JAW07502.1 JRES01000561 KNC30197.1 KQ971359 EFA08638.2 DS234986 EEB09842.1 KQ982268 KYQ58474.1 GFDL01014970 JAV20075.1 KK853598 KDR06420.1 GDIQ01108127 JAL43599.1 GDIQ01053748 JAN40989.1 GDIQ01174495 JAK77230.1 GDIQ01034999 JAN59738.1 ABLF02026514 GAMC01011438 JAB95117.1 GDHC01009044 JAQ09585.1 GDHC01011513 JAQ07116.1 GBHO01044682 JAF98921.1 DS231844 EDS35735.1 CVRI01000010 CRK88797.1 GDIQ01010006 JAN84731.1 GDIP01221463 JAJ01939.1 GFXV01006019 MBW17824.1 GBXI01000276 JAD14016.1 GDHF01025007 JAI27307.1 GAKP01006140 JAC52812.1 CCAG010003896 GGMS01012803 MBY82006.1 GGFL01004640 MBW68818.1 GGFJ01001756 MBW50897.1 GGFL01004641 MBW68819.1 GGFJ01001757 MBW50898.1 GGFJ01001758 MBW50899.1 ADMH02002133 ETN58440.1 GGFK01007065 MBW40386.1 GGFK01007247 MBW40568.1 KQ414663 KOC65372.1 AAAB01008839 EAA05843.4 APCN01004553 GL732762 EFX65117.1 ATLV01014994 KE524999 KFB39803.1 AXCM01000251 HACA01007592 CDW24953.1 CH933807 EDW13108.2 CH902624 EDV33511.1 CH963920 EDW77706.1 AXCN02000868
AGBW02009499 OWR50682.1 RSAL01000118 RVE46764.1 KZ149936 PZC77143.1 AGBW02010450 OWR48487.1 KQ434769 KZC03787.1 NNAY01001730 OXU23104.1 KZ288311 PBC28511.1 GEZM01085172 GEZM01085171 GEZM01085170 JAV60155.1 APGK01035628 APGK01035629 APGK01035630 KB740928 ENN77884.1 GL445930 EFN88780.1 LBMM01004251 KMQ92660.1 GEBQ01008946 JAT31031.1 GL888058 EGI68546.1 GECZ01011759 JAS58010.1 GBBI01002988 JAC15724.1 NEVH01016943 PNF25173.1 GL762091 EFZ22265.1 ADTU01008085 ADTU01008086 ADTU01008087 ADTU01008088 GECU01001857 JAT05850.1 GEDC01021045 JAS16253.1 KQ981604 KYN39719.1 GEDC01025130 JAS12168.1 GDKW01000777 JAI55818.1 KB632064 ERL88468.1 GECL01002690 JAP03434.1 KQ979488 KYN21228.1 KQ976710 KYM77038.1 KK107088 QOIP01000008 EZA59800.1 RLU19525.1 GFTR01008584 JAW07842.1 KQ977133 KYN05487.1 CH477305 EJY57536.1 JXUM01092462 KQ563989 KXJ72968.1 JXUM01067297 KQ562443 KXJ75905.1 GFTR01008924 JAW07502.1 JRES01000561 KNC30197.1 KQ971359 EFA08638.2 DS234986 EEB09842.1 KQ982268 KYQ58474.1 GFDL01014970 JAV20075.1 KK853598 KDR06420.1 GDIQ01108127 JAL43599.1 GDIQ01053748 JAN40989.1 GDIQ01174495 JAK77230.1 GDIQ01034999 JAN59738.1 ABLF02026514 GAMC01011438 JAB95117.1 GDHC01009044 JAQ09585.1 GDHC01011513 JAQ07116.1 GBHO01044682 JAF98921.1 DS231844 EDS35735.1 CVRI01000010 CRK88797.1 GDIQ01010006 JAN84731.1 GDIP01221463 JAJ01939.1 GFXV01006019 MBW17824.1 GBXI01000276 JAD14016.1 GDHF01025007 JAI27307.1 GAKP01006140 JAC52812.1 CCAG010003896 GGMS01012803 MBY82006.1 GGFL01004640 MBW68818.1 GGFJ01001756 MBW50897.1 GGFL01004641 MBW68819.1 GGFJ01001757 MBW50898.1 GGFJ01001758 MBW50899.1 ADMH02002133 ETN58440.1 GGFK01007065 MBW40386.1 GGFK01007247 MBW40568.1 KQ414663 KOC65372.1 AAAB01008839 EAA05843.4 APCN01004553 GL732762 EFX65117.1 ATLV01014994 KE524999 KFB39803.1 AXCM01000251 HACA01007592 CDW24953.1 CH933807 EDW13108.2 CH902624 EDV33511.1 CH963920 EDW77706.1 AXCN02000868
Proteomes
UP000005204
UP000053240
UP000007151
UP000283053
UP000192223
UP000076502
+ More
UP000005203 UP000215335 UP000242457 UP000002358 UP000019118 UP000008237 UP000036403 UP000007755 UP000235965 UP000005205 UP000078541 UP000030742 UP000078492 UP000078540 UP000053097 UP000279307 UP000078542 UP000008820 UP000069940 UP000249989 UP000037069 UP000095300 UP000007266 UP000009046 UP000075809 UP000095301 UP000027135 UP000007819 UP000002320 UP000183832 UP000078200 UP000092444 UP000092443 UP000092445 UP000091820 UP000000673 UP000053825 UP000075882 UP000007062 UP000069272 UP000079169 UP000076407 UP000075840 UP000075884 UP000075900 UP000000305 UP000075903 UP000030765 UP000075902 UP000075883 UP000075885 UP000009192 UP000007801 UP000007798 UP000075886 UP000075881 UP000075920
UP000005203 UP000215335 UP000242457 UP000002358 UP000019118 UP000008237 UP000036403 UP000007755 UP000235965 UP000005205 UP000078541 UP000030742 UP000078492 UP000078540 UP000053097 UP000279307 UP000078542 UP000008820 UP000069940 UP000249989 UP000037069 UP000095300 UP000007266 UP000009046 UP000075809 UP000095301 UP000027135 UP000007819 UP000002320 UP000183832 UP000078200 UP000092444 UP000092443 UP000092445 UP000091820 UP000000673 UP000053825 UP000075882 UP000007062 UP000069272 UP000079169 UP000076407 UP000075840 UP000075884 UP000075900 UP000000305 UP000075903 UP000030765 UP000075902 UP000075883 UP000075885 UP000009192 UP000007801 UP000007798 UP000075886 UP000075881 UP000075920
PRIDE
Pfam
Interpro
IPR007644
RNA_pol_bsu_protrusion
+ More
IPR037034 RNA_pol_Rpb2_2_sf
IPR014724 RNA_pol_RPB2_OB-fold
IPR037033 DNA-dir_RNAP_su2_hyb_sf
IPR007645 RNA_pol_Rpb2_3
IPR015712 DNA-dir_RNA_pol_su2
IPR007121 RNA_pol_bsu_CS
IPR009674 Rpa2_dom_4
IPR007642 RNA_pol_Rpb2_2
IPR007120 DNA-dir_RNAP_su2_dom
IPR007641 RNA_pol_Rpb2_7
IPR022083 KBP
IPR037034 RNA_pol_Rpb2_2_sf
IPR014724 RNA_pol_RPB2_OB-fold
IPR037033 DNA-dir_RNAP_su2_hyb_sf
IPR007645 RNA_pol_Rpb2_3
IPR015712 DNA-dir_RNA_pol_su2
IPR007121 RNA_pol_bsu_CS
IPR009674 Rpa2_dom_4
IPR007642 RNA_pol_Rpb2_2
IPR007120 DNA-dir_RNAP_su2_dom
IPR007641 RNA_pol_Rpb2_7
IPR022083 KBP
Gene 3D
ProteinModelPortal
H9JMQ1
A0A0N1ICC8
A0A212FAC3
A0A3S2NX40
A0A2W1BZB1
A0A212F425
+ More
A0A1W4XHB8 A0A154NVW8 A0A088A2A1 A0A232EXH0 A0A2A3EBT9 A0A1Y1KKJ0 K7IWJ3 N6TC52 E2B687 A0A0J7NJI8 A0A1B6M535 F4WB73 A0A1B6G6E8 A0A023F422 A0A2J7Q9B5 E9IAZ0 A0A158P3I7 A0A1B6K312 A0A1B6CSM5 A0A151JXF7 A0A1B6CFG7 A0A0P4VRX0 U4U813 A0A0V0G7L3 A0A151J8Q7 A0A151HZI2 A0A026WUS5 A0A224XGY9 A0A151IL56 J9E9J9 A0A182GW05 A0A182GJ47 A0A224XFY6 A0A0L0CDA1 A0A1I8QCQ3 D6WVZ3 E0V8Y6 A0A151XDM1 A0A1Q3EXN9 A0A1I8M669 A0A067QQ90 A0A0P5QZ94 A0A0N8E7P0 A0A0P5L5Y1 A0A0P6GD02 J9K6G2 W8B904 A0A146LR30 A0A146LJQ4 A0A0A9VR72 B0W5T2 A0A1J1HL88 A0A0P6IJK5 A0A0P4ZUJ3 A0A2H8TU91 A0A0A1XSL8 A0A0K8UKU8 A0A034WES4 A0A1A9V0X0 A0A1B0FD37 A0A1A9YSF8 A0A1A9ZQR0 A0A2S2QW78 A0A1A9WUR2 A0A2M4CVF7 A0A2M4BD15 A0A2M4CU57 A0A2M4BDL2 A0A2M4BCX8 W5J6H6 A0A2M4AHX0 A0A2M4AIY3 A0A0L7R3C5 A0A182L5Q9 Q7QG74 A0A182F4E3 A0A3Q0ITX4 A0A182X7Q7 A0A182IE12 A0A182NE81 A0A182RGH5 E9HT05 A0A182VG37 A0A084VPA9 A0A182TZ99 A0A182MM49 A0A0K2TFZ6 A0A182P955 B4KFJ3 B3MUI2 B4N0U3 A0A182Q1G0 A0A182JYE9 A0A182VV21
A0A1W4XHB8 A0A154NVW8 A0A088A2A1 A0A232EXH0 A0A2A3EBT9 A0A1Y1KKJ0 K7IWJ3 N6TC52 E2B687 A0A0J7NJI8 A0A1B6M535 F4WB73 A0A1B6G6E8 A0A023F422 A0A2J7Q9B5 E9IAZ0 A0A158P3I7 A0A1B6K312 A0A1B6CSM5 A0A151JXF7 A0A1B6CFG7 A0A0P4VRX0 U4U813 A0A0V0G7L3 A0A151J8Q7 A0A151HZI2 A0A026WUS5 A0A224XGY9 A0A151IL56 J9E9J9 A0A182GW05 A0A182GJ47 A0A224XFY6 A0A0L0CDA1 A0A1I8QCQ3 D6WVZ3 E0V8Y6 A0A151XDM1 A0A1Q3EXN9 A0A1I8M669 A0A067QQ90 A0A0P5QZ94 A0A0N8E7P0 A0A0P5L5Y1 A0A0P6GD02 J9K6G2 W8B904 A0A146LR30 A0A146LJQ4 A0A0A9VR72 B0W5T2 A0A1J1HL88 A0A0P6IJK5 A0A0P4ZUJ3 A0A2H8TU91 A0A0A1XSL8 A0A0K8UKU8 A0A034WES4 A0A1A9V0X0 A0A1B0FD37 A0A1A9YSF8 A0A1A9ZQR0 A0A2S2QW78 A0A1A9WUR2 A0A2M4CVF7 A0A2M4BD15 A0A2M4CU57 A0A2M4BDL2 A0A2M4BCX8 W5J6H6 A0A2M4AHX0 A0A2M4AIY3 A0A0L7R3C5 A0A182L5Q9 Q7QG74 A0A182F4E3 A0A3Q0ITX4 A0A182X7Q7 A0A182IE12 A0A182NE81 A0A182RGH5 E9HT05 A0A182VG37 A0A084VPA9 A0A182TZ99 A0A182MM49 A0A0K2TFZ6 A0A182P955 B4KFJ3 B3MUI2 B4N0U3 A0A182Q1G0 A0A182JYE9 A0A182VV21
PDB
6HLS
E-value=6.068e-151,
Score=1373
Ontologies
GO
PANTHER
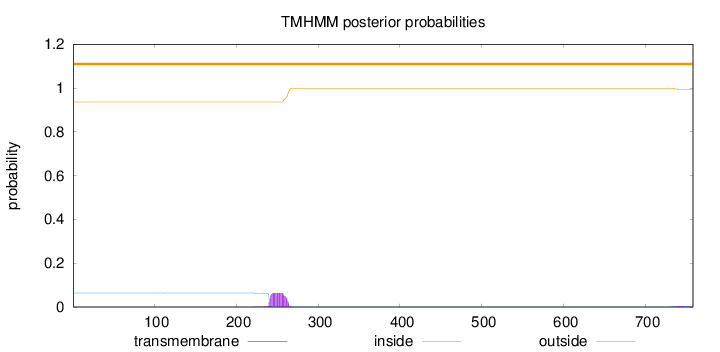
Topology
Length:
758
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.45956
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.06361
outside
1 - 758
Population Genetic Test Statistics
Pi
230.40007
Theta
176.378838
Tajima's D
0.927695
CLR
0.236243
CSRT
0.638518074096295
Interpretation
Uncertain
