Pre Gene Modal
BGIBMGA010740
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_facilitated_trehalose_transporter_Tret1-2_homolog_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.95
Sequence
CDS
ATGGACAGACCTACTATGCACATGCTCTCCGTCGAGGAGAAGGGCGGAAAAACACAAAGGACCAATCAGTATATTGCTGCAATTGCTGCTGCCATTGGAGCAGTAGCCGCCGGCACCATCCTGGCTTGGACGTCTCCCGTTCTACCCCAACTGGAACCTTGGAAGGACGATGATAATCAAACTCTCCCAAACAACACGACGCCAGCACCGCTTGACTACCGAAAGAACGCCACCAATTCTAGCCAACTGATTAACTCCCTCGGCGAGCCCGCTGATTTCCTTCTGGACACGAGTACAAAATCTCTGGTGTCTTCTATCATGGCTATCGGCGCGGCTATAGCGGCGCTCCCTGTCGGATTCTCGGCCCAACGCTTCGGACGCCGGGTCACTATACTGATCCTCGTTATACCCCTAATGATCAACTGGCTGCTGACGATCTTCGCCAACGGGGCCGGTATGCTGATCGCTGCTAGATTCTTCGCGGGTCTTGCTACTGGTGGAATGTGCGTGGCGGCTCCCCTGTACATCGGGGAGATAGCGGAGACCGCGATCCGTGGCGCTTTGGGCTCGTTCTTCCAGCTTTTCATTACTGTCGGCATCCTATTCACGTTCGTCGTCGGCGGCTGGACGCATTGGAAGACCCTGTCCATCATTAACGCAGCGGTGCCTTTATTGCTTGTTGCTGTATTTTGGTGGATGCCGGAGACGCCGCAGTACCTACTGTCGTGTGGGCGACGGCGCGAGGCCGAGCAGTCGCTGCGCTGGTTGCGGGGCCCGCACGCCGACCTTGCCGCCGAGTTGGACGAGATGCAGAAAGACGTGGAGGCGTCGGGGCGCGCGCGGGGCGGGGTGCTGGCGCTGACGCGGGGCGGGGCCACTCGCCGCGCCATGCTGTGCGGGCTCGGCCTCATGCTCTTCCAGCAGTTCAGCGGCGTCAACGCAGTCATCTTCTACACCGTGCAGATCTTCCGTTCCGCCGGCTCCGACATACCGTCCGTGATCGCCACCATCATCGTGGGCGTGGTCCAGACCATCTCAACCGTCGGGTCCTCGCTGCTCATCGAGCGAGCCGGCCGGAGGATCCTCTTGCTGCAGAGCTGCATCATCATGGGGCTGTCCCTGGTCGTATTGGGAACTTACTTCAAGTTACAGAGGGACGGTGTCGCGTTAGCCGTCGTCGGGTGGATCCCGCTGTTGTGTTTGGTGCTCTTCATCATTTCGTTTTCGATGGGATTCGGCCCGATACCCTGGACGATGATGTCAGAACTGTTCCCCGCCGAGATCAGGGGGACGGCCGCGGGGATCACGGTCGTAGTGAACTGGGTCCTAGTGTTCATTGTTACTCTCTGCTTCCCGCTGATGATGGACGCGTTGGGGATCTACAGCTGCTTCTGGTTCTTCGCTGCCTTCATGCTGGTCTGCGTGTTCTTCGTGTCCTTCCTGATCCCCGAGACGAAGGGGAAGTCTTTATCGCAGATCCAGGCCATCCTGGCCGGAAAGAGGATCTGA
Protein
MDRPTMHMLSVEEKGGKTQRTNQYIAAIAAAIGAVAAGTILAWTSPVLPQLEPWKDDDNQTLPNNTTPAPLDYRKNATNSSQLINSLGEPADFLLDTSTKSLVSSIMAIGAAIAALPVGFSAQRFGRRVTILILVIPLMINWLLTIFANGAGMLIAARFFAGLATGGMCVAAPLYIGEIAETAIRGALGSFFQLFITVGILFTFVVGGWTHWKTLSIINAAVPLLLVAVFWWMPETPQYLLSCGRRREAEQSLRWLRGPHADLAAELDEMQKDVEASGRARGGVLALTRGGATRRAMLCGLGLMLFQQFSGVNAVIFYTVQIFRSAGSDIPSVIATIIVGVVQTISTVGSSLLIERAGRRILLLQSCIIMGLSLVVLGTYFKLQRDGVALAVVGWIPLLCLVLFIISFSMGFGPIPWTMMSELFPAEIRGTAAGITVVVNWVLVFIVTLCFPLMMDALGIYSCFWFFAAFMLVCVFFVSFLIPETKGKSLSQIQAILAGKRI
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JMI7
A0A2A4JPP7
A0A2W1BU14
S4PEN4
A0A212F416
A0A3S2NG77
+ More
A0A2H1VB69 A0A0N1IGQ6 A0A194PLS6 A0A182P8W6 A0A182R879 A0A2M3YZG0 A0A182K847 A0A182QJ59 A0A182MVH7 A0A182VS59 A0A182WW12 F5HJ72 A0A182UFU4 A0A182KX78 A0A2M4BLQ8 A0A182FKZ3 A0A2M4A0Z4 A0A2M4A1V1 A0A2M4A607 Q7PCM5 A0A182HJY9 A0A182NL12 W5J2M6 A0A1S4FQN7 A0A182JL03 A0A084VDW2 A0A1Q3FK39 A0A1Q3FK71 A0A182YLM9 A0A2M4CLI7 A0A182T8Z5 U5EUX1 A0A336LI37 A0A182GL68 B0WZW6 A0A1Y1KQ78 A0A2P8YT21 B4LXB4 A0A0Q9WS19 A0A2M4CMG9 B4NJT8 A0A1W4UJE8 A0A3B0JHR8 A0A1W4UYK2 A0A182UZF0 B3LWC6 B5DW12 B4G347 A0A0N8P1N8 I5AP16 B4KAB2 A0A0Q9WYV8 A0A0Q9WYV1 A0A1L8DIV5 A0A0L0CIX6 A0A1L8DIY0 B4JH51 A0A1I8NPR6 A0A1I8NPR8 B4I486 B4QXL3 B3NYI1 A0A1I8MYG4 B4PVJ2 A0A0R1E4G7 T1PG78 A0A0K8VUC9 A0A0K8UKG0 A0A1W4XL19 A0A034VDB8 A0A034V8B7 A0A0M3QYJ6 A0A0A1XNV7 Q8T0T6 A0A2J7PIQ0 A8JQU1 A0A1J1IMH8 E9IQR5 U5EQL0 W8ATJ1 A0A1B6L0T0 B4ML33 A0A1B6MT94 A0A0A1WG49 A0A0A1WTN4 Q9VU17 Q8IQH6 A0A034VPA7 A0A0J9UK27 A0A0J9RVP2 B4HG70
A0A2H1VB69 A0A0N1IGQ6 A0A194PLS6 A0A182P8W6 A0A182R879 A0A2M3YZG0 A0A182K847 A0A182QJ59 A0A182MVH7 A0A182VS59 A0A182WW12 F5HJ72 A0A182UFU4 A0A182KX78 A0A2M4BLQ8 A0A182FKZ3 A0A2M4A0Z4 A0A2M4A1V1 A0A2M4A607 Q7PCM5 A0A182HJY9 A0A182NL12 W5J2M6 A0A1S4FQN7 A0A182JL03 A0A084VDW2 A0A1Q3FK39 A0A1Q3FK71 A0A182YLM9 A0A2M4CLI7 A0A182T8Z5 U5EUX1 A0A336LI37 A0A182GL68 B0WZW6 A0A1Y1KQ78 A0A2P8YT21 B4LXB4 A0A0Q9WS19 A0A2M4CMG9 B4NJT8 A0A1W4UJE8 A0A3B0JHR8 A0A1W4UYK2 A0A182UZF0 B3LWC6 B5DW12 B4G347 A0A0N8P1N8 I5AP16 B4KAB2 A0A0Q9WYV8 A0A0Q9WYV1 A0A1L8DIV5 A0A0L0CIX6 A0A1L8DIY0 B4JH51 A0A1I8NPR6 A0A1I8NPR8 B4I486 B4QXL3 B3NYI1 A0A1I8MYG4 B4PVJ2 A0A0R1E4G7 T1PG78 A0A0K8VUC9 A0A0K8UKG0 A0A1W4XL19 A0A034VDB8 A0A034V8B7 A0A0M3QYJ6 A0A0A1XNV7 Q8T0T6 A0A2J7PIQ0 A8JQU1 A0A1J1IMH8 E9IQR5 U5EQL0 W8ATJ1 A0A1B6L0T0 B4ML33 A0A1B6MT94 A0A0A1WG49 A0A0A1WTN4 Q9VU17 Q8IQH6 A0A034VPA7 A0A0J9UK27 A0A0J9RVP2 B4HG70
Pubmed
19121390
28756777
23622113
22118469
26354079
12364791
+ More
14747013 17210077 20966253 20920257 23761445 17510324 24438588 25244985 26483478 28004739 29403074 17994087 15632085 26108605 25315136 17550304 25348373 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 24495485 22936249
14747013 17210077 20966253 20920257 23761445 17510324 24438588 25244985 26483478 28004739 29403074 17994087 15632085 26108605 25315136 17550304 25348373 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 24495485 22936249
EMBL
BABH01016704
BABH01016705
NWSH01000916
PCG73554.1
KZ149936
PZC77145.1
+ More
GAIX01004412 JAA88148.1 AGBW02010450 OWR48486.1 RSAL01000118 RVE46763.1 ODYU01001615 SOQ38089.1 KQ459910 KPJ19331.1 KQ459601 KPI93689.1 GGFM01000885 MBW21636.1 AXCN02000090 AXCM01000233 AAAB01008820 EGK96332.1 EGK96333.1 EGK96334.1 GGFJ01004878 MBW54019.1 GGFK01001165 MBW34486.1 GGFK01001408 MBW34729.1 GGFK01002916 MBW36237.1 EAA05397.6 APCN01002224 ADMH02002130 ETN58527.1 ATLV01011810 KE524723 KFB36156.1 GFDL01007161 JAV27884.1 GFDL01007076 JAV27969.1 GGFL01002019 MBW66197.1 GANO01002135 JAB57736.1 UFQT01000014 SSX17682.1 JXUM01071357 JXUM01071358 JXUM01071359 JXUM01071360 KQ562660 KXJ75377.1 DS232219 EDS37791.1 GEZM01078913 JAV62581.1 PYGN01000377 PSN47391.1 CH940650 EDW67792.1 KRF83470.1 GGFL01002362 MBW66540.1 CH964272 EDW85050.2 OUUW01000005 SPP80253.1 CH902617 EDV43759.1 CM000070 EDY68099.1 CH479179 EDW24242.1 KPU80444.1 EIM52701.1 CH933806 EDW14599.1 KRG01100.1 KRG01099.1 GFDF01007787 JAV06297.1 JRES01000423 KNC31409.1 GFDF01007790 JAV06294.1 CH916369 EDV92742.1 CH480821 EDW55029.1 CM000364 EDX11799.1 CH954181 EDV47962.1 CM000160 EDW97801.1 KRK03977.1 KA647145 AFP61774.1 GDHF01009846 JAI42468.1 GDHF01025504 JAI26810.1 GAKP01019399 JAC39553.1 GAKP01019401 GAKP01019400 JAC39551.1 CP012526 ALC47793.1 GBXI01001611 JAD12681.1 AE014297 AY069071 AAF51943.2 AAL39216.1 NEVH01025074 PNF16189.1 ABW08606.1 CVRI01000055 CRL01368.1 GL764900 EFZ17072.1 GANO01004191 JAB55680.1 GAMC01014390 JAB92165.1 GEBQ01022823 JAT17154.1 CH963847 EDW73091.2 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 GBXI01016924 JAC97367.1 GBXI01012080 JAD02212.1 AE014296 AY119564 AAF49874.1 AAM50218.1 AAN11863.1 AAN11864.1 GAKP01015559 GAKP01015558 GAKP01015557 JAC43393.1 CM002912 KMY99310.1 KMY99309.1 CH480815 EDW41316.1
GAIX01004412 JAA88148.1 AGBW02010450 OWR48486.1 RSAL01000118 RVE46763.1 ODYU01001615 SOQ38089.1 KQ459910 KPJ19331.1 KQ459601 KPI93689.1 GGFM01000885 MBW21636.1 AXCN02000090 AXCM01000233 AAAB01008820 EGK96332.1 EGK96333.1 EGK96334.1 GGFJ01004878 MBW54019.1 GGFK01001165 MBW34486.1 GGFK01001408 MBW34729.1 GGFK01002916 MBW36237.1 EAA05397.6 APCN01002224 ADMH02002130 ETN58527.1 ATLV01011810 KE524723 KFB36156.1 GFDL01007161 JAV27884.1 GFDL01007076 JAV27969.1 GGFL01002019 MBW66197.1 GANO01002135 JAB57736.1 UFQT01000014 SSX17682.1 JXUM01071357 JXUM01071358 JXUM01071359 JXUM01071360 KQ562660 KXJ75377.1 DS232219 EDS37791.1 GEZM01078913 JAV62581.1 PYGN01000377 PSN47391.1 CH940650 EDW67792.1 KRF83470.1 GGFL01002362 MBW66540.1 CH964272 EDW85050.2 OUUW01000005 SPP80253.1 CH902617 EDV43759.1 CM000070 EDY68099.1 CH479179 EDW24242.1 KPU80444.1 EIM52701.1 CH933806 EDW14599.1 KRG01100.1 KRG01099.1 GFDF01007787 JAV06297.1 JRES01000423 KNC31409.1 GFDF01007790 JAV06294.1 CH916369 EDV92742.1 CH480821 EDW55029.1 CM000364 EDX11799.1 CH954181 EDV47962.1 CM000160 EDW97801.1 KRK03977.1 KA647145 AFP61774.1 GDHF01009846 JAI42468.1 GDHF01025504 JAI26810.1 GAKP01019399 JAC39553.1 GAKP01019401 GAKP01019400 JAC39551.1 CP012526 ALC47793.1 GBXI01001611 JAD12681.1 AE014297 AY069071 AAF51943.2 AAL39216.1 NEVH01025074 PNF16189.1 ABW08606.1 CVRI01000055 CRL01368.1 GL764900 EFZ17072.1 GANO01004191 JAB55680.1 GAMC01014390 JAB92165.1 GEBQ01022823 JAT17154.1 CH963847 EDW73091.2 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 GBXI01016924 JAC97367.1 GBXI01012080 JAD02212.1 AE014296 AY119564 AAF49874.1 AAM50218.1 AAN11863.1 AAN11864.1 GAKP01015559 GAKP01015558 GAKP01015557 JAC43393.1 CM002912 KMY99310.1 KMY99309.1 CH480815 EDW41316.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000075885 UP000075900 UP000075881 UP000075886 UP000075883 UP000075920 UP000076407 UP000007062 UP000075902 UP000075882 UP000069272 UP000075840 UP000075884 UP000000673 UP000075880 UP000030765 UP000076408 UP000075901 UP000069940 UP000249989 UP000002320 UP000245037 UP000008792 UP000007798 UP000192221 UP000268350 UP000075903 UP000007801 UP000001819 UP000008744 UP000009192 UP000037069 UP000001070 UP000095300 UP000001292 UP000000304 UP000008711 UP000095301 UP000002282 UP000192223 UP000092553 UP000000803 UP000235965 UP000183832
UP000075885 UP000075900 UP000075881 UP000075886 UP000075883 UP000075920 UP000076407 UP000007062 UP000075902 UP000075882 UP000069272 UP000075840 UP000075884 UP000000673 UP000075880 UP000030765 UP000076408 UP000075901 UP000069940 UP000249989 UP000002320 UP000245037 UP000008792 UP000007798 UP000192221 UP000268350 UP000075903 UP000007801 UP000001819 UP000008744 UP000009192 UP000037069 UP000001070 UP000095300 UP000001292 UP000000304 UP000008711 UP000095301 UP000002282 UP000192223 UP000092553 UP000000803 UP000235965 UP000183832
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JMI7
A0A2A4JPP7
A0A2W1BU14
S4PEN4
A0A212F416
A0A3S2NG77
+ More
A0A2H1VB69 A0A0N1IGQ6 A0A194PLS6 A0A182P8W6 A0A182R879 A0A2M3YZG0 A0A182K847 A0A182QJ59 A0A182MVH7 A0A182VS59 A0A182WW12 F5HJ72 A0A182UFU4 A0A182KX78 A0A2M4BLQ8 A0A182FKZ3 A0A2M4A0Z4 A0A2M4A1V1 A0A2M4A607 Q7PCM5 A0A182HJY9 A0A182NL12 W5J2M6 A0A1S4FQN7 A0A182JL03 A0A084VDW2 A0A1Q3FK39 A0A1Q3FK71 A0A182YLM9 A0A2M4CLI7 A0A182T8Z5 U5EUX1 A0A336LI37 A0A182GL68 B0WZW6 A0A1Y1KQ78 A0A2P8YT21 B4LXB4 A0A0Q9WS19 A0A2M4CMG9 B4NJT8 A0A1W4UJE8 A0A3B0JHR8 A0A1W4UYK2 A0A182UZF0 B3LWC6 B5DW12 B4G347 A0A0N8P1N8 I5AP16 B4KAB2 A0A0Q9WYV8 A0A0Q9WYV1 A0A1L8DIV5 A0A0L0CIX6 A0A1L8DIY0 B4JH51 A0A1I8NPR6 A0A1I8NPR8 B4I486 B4QXL3 B3NYI1 A0A1I8MYG4 B4PVJ2 A0A0R1E4G7 T1PG78 A0A0K8VUC9 A0A0K8UKG0 A0A1W4XL19 A0A034VDB8 A0A034V8B7 A0A0M3QYJ6 A0A0A1XNV7 Q8T0T6 A0A2J7PIQ0 A8JQU1 A0A1J1IMH8 E9IQR5 U5EQL0 W8ATJ1 A0A1B6L0T0 B4ML33 A0A1B6MT94 A0A0A1WG49 A0A0A1WTN4 Q9VU17 Q8IQH6 A0A034VPA7 A0A0J9UK27 A0A0J9RVP2 B4HG70
A0A2H1VB69 A0A0N1IGQ6 A0A194PLS6 A0A182P8W6 A0A182R879 A0A2M3YZG0 A0A182K847 A0A182QJ59 A0A182MVH7 A0A182VS59 A0A182WW12 F5HJ72 A0A182UFU4 A0A182KX78 A0A2M4BLQ8 A0A182FKZ3 A0A2M4A0Z4 A0A2M4A1V1 A0A2M4A607 Q7PCM5 A0A182HJY9 A0A182NL12 W5J2M6 A0A1S4FQN7 A0A182JL03 A0A084VDW2 A0A1Q3FK39 A0A1Q3FK71 A0A182YLM9 A0A2M4CLI7 A0A182T8Z5 U5EUX1 A0A336LI37 A0A182GL68 B0WZW6 A0A1Y1KQ78 A0A2P8YT21 B4LXB4 A0A0Q9WS19 A0A2M4CMG9 B4NJT8 A0A1W4UJE8 A0A3B0JHR8 A0A1W4UYK2 A0A182UZF0 B3LWC6 B5DW12 B4G347 A0A0N8P1N8 I5AP16 B4KAB2 A0A0Q9WYV8 A0A0Q9WYV1 A0A1L8DIV5 A0A0L0CIX6 A0A1L8DIY0 B4JH51 A0A1I8NPR6 A0A1I8NPR8 B4I486 B4QXL3 B3NYI1 A0A1I8MYG4 B4PVJ2 A0A0R1E4G7 T1PG78 A0A0K8VUC9 A0A0K8UKG0 A0A1W4XL19 A0A034VDB8 A0A034V8B7 A0A0M3QYJ6 A0A0A1XNV7 Q8T0T6 A0A2J7PIQ0 A8JQU1 A0A1J1IMH8 E9IQR5 U5EQL0 W8ATJ1 A0A1B6L0T0 B4ML33 A0A1B6MT94 A0A0A1WG49 A0A0A1WTN4 Q9VU17 Q8IQH6 A0A034VPA7 A0A0J9UK27 A0A0J9RVP2 B4HG70
PDB
4LDS
E-value=2.31162e-48,
Score=486
Ontologies
GO
Topology
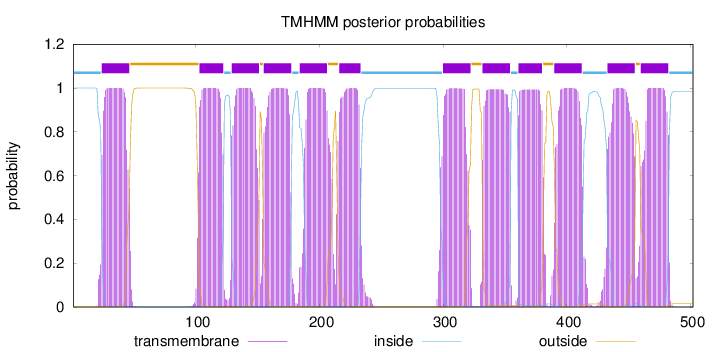
Length:
502
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
261.30341
Exp number, first 60 AAs:
22.1914
Total prob of N-in:
0.99975
POSSIBLE N-term signal
sequence
inside
1 - 23
TMhelix
24 - 46
outside
47 - 102
TMhelix
103 - 122
inside
123 - 128
TMhelix
129 - 151
outside
152 - 154
TMhelix
155 - 177
inside
178 - 183
TMhelix
184 - 206
outside
207 - 215
TMhelix
216 - 233
inside
234 - 299
TMhelix
300 - 322
outside
323 - 331
TMhelix
332 - 354
inside
355 - 360
TMhelix
361 - 380
outside
381 - 389
TMhelix
390 - 412
inside
413 - 432
TMhelix
433 - 455
outside
456 - 459
TMhelix
460 - 482
inside
483 - 502
Population Genetic Test Statistics
Pi
228.291346
Theta
160.93984
Tajima's D
1.538679
CLR
0.071962
CSRT
0.795310234488276
Interpretation
Uncertain
