Gene
KWMTBOMO15540
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.4
Sequence
CDS
ATGTTCTTTGTGGGATATGTTAAGTCTCTAGTCTACGCCGATAAGCCCGAAACGATTGACCACTTGGAGGACAACATTCGCCGCGCTATTGCCGACAGGGCTCTGAAACTGCAATCCTTTTTACTGGTGGTAGGACCTCTTGTCAGGGCGGTTCGCGGGTACCGCACCGTCTCGTTCGAGGCGGCGAGCGTACTCGCCGGGACGCCTCCCTGGGACTTGGAGGCGGAGGCGCTCGCTGCGGATTACGCGTGGCGATGCGATCTCCGCTCCAGGGGGGAGCCGCGTCCCGGCGCGGCGGAAGTTCGAGCGCGGAAGCTTCAATCTCGGCGTGTCGTACTCGAGGCGTGGTCTCGCCGCCTGGCGGACCCCGCCTACGGGCGGCGGACCGTCGAGGCGATCCGCCCGGTCCTCTCGGGCTGGGTGAATCGCGACCGAGAACGTCTCACCTTCCGAGCGACGCAGGTGCTAACGGGACACGGCTGTTTCGGTCGCTACCTGCACCTCATCGCCCGGAGGGAGCCGACGCCGAAGTGCCACCACTGCAGTGGTTGCAACGAGGACACAGCGGAGCACACGCTTGCGTACTGCCCCGCTTTCGCGGAGCAGCGCCGCGTCCTCGTTGCAAAAATAGGACCGGACTTGTCGTTTCCGACCGTCGTGGCTACGATGCTCGGCAGCGTCGAGTCCTGGCAGGCGATGCTCGACTTCTCCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAACGGGAGAGGGAGAGCTCTTCTCCCCTCTCGTCGCCGTGCCGCCGTCGCTGA
Protein
MFFVGYVKSLVYADKPETIDHLEDNIRRAIADRALKLQSFLLVVGPLVRAVRGYRTVSFEAASVLAGTPPWDLEAEALAADYAWRCDLRSRGEPRPGAAEVRARKLQSRRVVLEAWSRRLADPAYGRRTVEAIRPVLSGWVNRDRERLTFRATQVLTGHGCFGRYLHLIARREPTPKCHHCSGCNEDTAEHTLAYCPAFAEQRRVLVAKIGPDLSFPTVVATMLGSVESWQAMLDFSESTISQKEAAERERESSSPLSSPCRRR
Summary
Uniprot
Q5KTM7
Q868Q4
A0A0J7KCI1
A0A2W1BDU5
A0A2W1B333
A0A2W1BSQ5
+ More
A0A0J7N1D9 Q8MY27 A0A2W1C3J5 A0A3L8DDS1 Q8MY33 Q8MY31 A0A0J7MZM4 A0A0J7KJB9 Q8MY35 A0A2H1WNZ5 A0A0J7N7H9 A0A0J7KS67 A0A3S2N998 A0A0J7KIY5 A0A0J7N7S8 A0A0J7MY07 A0A0J7JXN3 O01419 A0A0J7KCQ3 A0A0J7KIW1 A0A0J7KJ07 A0A0J7NEG9 A0A194R5D3 A0A0J7KJG6 A0A0J7KYD2 A0A0J7KM81 A0A0J7KQK3 A0A0J7JZF7 A0A0J7JZ13 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KJH4 A0A0J7KHX7 A0A3L8DDV9 A0A0J7KCR7 A0A3L8D2G3 J9KZ24 A0A0Q9WRU0 J9KLZ3 A0A0J7MX22
A0A0J7N1D9 Q8MY27 A0A2W1C3J5 A0A3L8DDS1 Q8MY33 Q8MY31 A0A0J7MZM4 A0A0J7KJB9 Q8MY35 A0A2H1WNZ5 A0A0J7N7H9 A0A0J7KS67 A0A3S2N998 A0A0J7KIY5 A0A0J7N7S8 A0A0J7MY07 A0A0J7JXN3 O01419 A0A0J7KCQ3 A0A0J7KIW1 A0A0J7KJ07 A0A0J7NEG9 A0A194R5D3 A0A0J7KJG6 A0A0J7KYD2 A0A0J7KM81 A0A0J7KQK3 A0A0J7JZF7 A0A0J7JZ13 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KJH4 A0A0J7KHX7 A0A3L8DDV9 A0A0J7KCR7 A0A3L8D2G3 J9KZ24 A0A0Q9WRU0 J9KLZ3 A0A0J7MX22
EMBL
AB126050
BAD86650.1
AB090825
BAC57926.1
LBMM01009501
KMQ88083.1
+ More
KZ150308 PZC71477.1 KZ150386 PZC71062.1 KZ149950 PZC76665.1 LBMM01012027 KMQ86470.1 AB078935 BAC06462.1 KZ149896 PZC78733.1 QOIP01000009 RLU18544.1 AB078930 BAC06454.1 AB078931 BAC06456.1 LBMM01012957 LBMM01012956 KMQ85900.1 KMQ85901.1 LBMM01006538 KMQ90523.1 AB078929 BAC06452.1 ODYU01009945 SOQ54696.1 LBMM01008861 KMQ88580.1 LBMM01003679 KMQ93242.1 RSAL01000481 RVE41577.1 LBMM01006954 KMQ90166.1 LBMM01008682 KMQ88705.1 LBMM01014132 KMQ85305.1 LBMM01024024 KMQ82631.1 D85594 BAA19776.1 LBMM01009494 KMQ88097.1 LBMM01006725 KMQ90373.1 LBMM01006662 KMQ90413.1 LBMM01006110 KMQ90925.1 KQ460685 KPJ12887.1 LBMM01006671 KMQ90407.1 LBMM01002052 KMQ95336.1 LBMM01005634 KMQ91349.1 LBMM01004272 KMQ92618.1 LBMM01019838 KMQ83439.1 LBMM01019839 KMQ83438.1 LBMM01005733 KMQ91252.1 LBMM01007036 KMQ90099.1 LBMM01006657 KMQ90417.1 LBMM01007088 KMQ90053.1 QOIP01000010 RLU18098.1 LBMM01009472 KMQ88117.1 QOIP01000031 RLU14697.1 ABLF02006132 CH963857 KRF98317.1 ABLF02023401 ABLF02029423 ABLF02060278 LBMM01014909 KMQ85000.1
KZ150308 PZC71477.1 KZ150386 PZC71062.1 KZ149950 PZC76665.1 LBMM01012027 KMQ86470.1 AB078935 BAC06462.1 KZ149896 PZC78733.1 QOIP01000009 RLU18544.1 AB078930 BAC06454.1 AB078931 BAC06456.1 LBMM01012957 LBMM01012956 KMQ85900.1 KMQ85901.1 LBMM01006538 KMQ90523.1 AB078929 BAC06452.1 ODYU01009945 SOQ54696.1 LBMM01008861 KMQ88580.1 LBMM01003679 KMQ93242.1 RSAL01000481 RVE41577.1 LBMM01006954 KMQ90166.1 LBMM01008682 KMQ88705.1 LBMM01014132 KMQ85305.1 LBMM01024024 KMQ82631.1 D85594 BAA19776.1 LBMM01009494 KMQ88097.1 LBMM01006725 KMQ90373.1 LBMM01006662 KMQ90413.1 LBMM01006110 KMQ90925.1 KQ460685 KPJ12887.1 LBMM01006671 KMQ90407.1 LBMM01002052 KMQ95336.1 LBMM01005634 KMQ91349.1 LBMM01004272 KMQ92618.1 LBMM01019838 KMQ83439.1 LBMM01019839 KMQ83438.1 LBMM01005733 KMQ91252.1 LBMM01007036 KMQ90099.1 LBMM01006657 KMQ90417.1 LBMM01007088 KMQ90053.1 QOIP01000010 RLU18098.1 LBMM01009472 KMQ88117.1 QOIP01000031 RLU14697.1 ABLF02006132 CH963857 KRF98317.1 ABLF02023401 ABLF02029423 ABLF02060278 LBMM01014909 KMQ85000.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q5KTM7
Q868Q4
A0A0J7KCI1
A0A2W1BDU5
A0A2W1B333
A0A2W1BSQ5
+ More
A0A0J7N1D9 Q8MY27 A0A2W1C3J5 A0A3L8DDS1 Q8MY33 Q8MY31 A0A0J7MZM4 A0A0J7KJB9 Q8MY35 A0A2H1WNZ5 A0A0J7N7H9 A0A0J7KS67 A0A3S2N998 A0A0J7KIY5 A0A0J7N7S8 A0A0J7MY07 A0A0J7JXN3 O01419 A0A0J7KCQ3 A0A0J7KIW1 A0A0J7KJ07 A0A0J7NEG9 A0A194R5D3 A0A0J7KJG6 A0A0J7KYD2 A0A0J7KM81 A0A0J7KQK3 A0A0J7JZF7 A0A0J7JZ13 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KJH4 A0A0J7KHX7 A0A3L8DDV9 A0A0J7KCR7 A0A3L8D2G3 J9KZ24 A0A0Q9WRU0 J9KLZ3 A0A0J7MX22
A0A0J7N1D9 Q8MY27 A0A2W1C3J5 A0A3L8DDS1 Q8MY33 Q8MY31 A0A0J7MZM4 A0A0J7KJB9 Q8MY35 A0A2H1WNZ5 A0A0J7N7H9 A0A0J7KS67 A0A3S2N998 A0A0J7KIY5 A0A0J7N7S8 A0A0J7MY07 A0A0J7JXN3 O01419 A0A0J7KCQ3 A0A0J7KIW1 A0A0J7KJ07 A0A0J7NEG9 A0A194R5D3 A0A0J7KJG6 A0A0J7KYD2 A0A0J7KM81 A0A0J7KQK3 A0A0J7JZF7 A0A0J7JZ13 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KJH4 A0A0J7KHX7 A0A3L8DDV9 A0A0J7KCR7 A0A3L8D2G3 J9KZ24 A0A0Q9WRU0 J9KLZ3 A0A0J7MX22
Ontologies
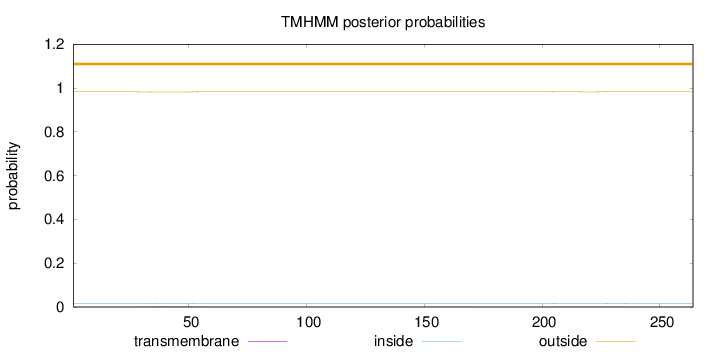
Topology
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05863
Exp number, first 60 AAs:
0.03089
Total prob of N-in:
0.01734
outside
1 - 264
Population Genetic Test Statistics
Pi
65.862297
Theta
67.348864
Tajima's D
0.006684
CLR
0
CSRT
0.369081545922704
Interpretation
Uncertain
