Gene
KWMTBOMO15536
Pre Gene Modal
BGIBMGA010743
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-2_homolog_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.982
Sequence
CDS
ATGTTTCTCGACCACTATGGCCTCCTTGGCATGTACTTCATATTGGACACCAACGGCAGTCCAATAGTCGACAGCATGAGGTTCCTACCCATTCTGGCGCTGACGCTGTTCATCATCTCATACTGCTTGGGCCTGGGTCCTCTGCCATGGGTTGTAATTGGAGAACTCTTCCCGATTGACGTGAAGGCGTTGGCTTCTCCAATAGCCACGGCCTTCTGCTGGCTGCTGTCCTTCTTGGTGACTAGATATTTCTACCTAGTAGCCGATATAGTTGGAATGGGTGTGGTCTTTCTCATCTTCGGAGTTTGCTGTATAGTAGCGTTCTTCTTCACATTCTTCGTGGTGCCGGAGACGAAGGGCAAGAGCTTCCAAGAGATCCAGGATATGCTGGCTGGGAAGGTGTCGCCATCGAAAACCGAGACAATTTAA
Protein
MFLDHYGLLGMYFILDTNGSPIVDSMRFLPILALTLFIISYCLGLGPLPWVVIGELFPIDVKALASPIATAFCWLLSFLVTRYFYLVADIVGMGVVFLIFGVCCIVAFFFTFFVVPETKGKSFQEIQDMLAGKVSPSKTETI
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JMJ0
H9JMI8
H9JMI9
A0A2H1VKV6
A0A2A4JNK2
A0A2A4JQ62
+ More
A0A2W1BVC4 A0A194PK39 A0A0L7LSC8 A0A2S2QIG2 J9JV25 A0A1L8DIL8 A0A1L8DIL5 A0A1L8DIJ3 A0A1L8DIK8 A0A212FL47 D7EK15 A0A182MM04 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182S579 A0A182VIR9 A0A182TSI6 V5I8R9 A0A1Y9H2R2 A0A182T9M2 A8CWB1 A0A2S2N8Q6 T1DE67 U5ESS0 A0A2H8TDY2 A0A1B0D4P3 A0A182HJZ0 A0A2M3ZFA3 A0A1Y0AWQ5 W5J543 A0A2M4A0H9 A0A2M4A0I2 E0W0A9 A0A182YLM9 A0A182QT48 A0A182VS58 T1DG93 A0A1Y1LWD0 A0A084VDW1 J3JXM2 N6SYK9 A0A2M4A1M5 A0A2M4BMJ9 A0A2M3ZF95 A0A182FKZ4 A0A1W4XAC6 A0A1L8DF28 U4UIC9 A0A1L8DF43 A0A139WCV1 A0A088ABY6 D7EK13 A0A2M3ZFG5 A0A034W685 A0A0J7L1I9 A0A1W4W7Z0 A0A1B0CZM5 A0A1J1IWB4 A0A1B6IS32 A0A195CMF0 A0A0C9RE51 E9IQR3 A0A1W4XJX1 A0A2A3E9H1 A0A336M0J9 A0A0J7NVW1 A0A0T6BH34 Q16SU5 W8BDP5 A0A0A1WDI7 B0WZW7 A0A0A1WV08 D4AHX1 W8B3Q4 A0A1B6L0T0 A0A1B6MT94 A0A182J8U1 A0A1B6M1S5 A0A2P8Z217 A0A1B6IVC7 A0A2P8Z215 A0A0K8W3N6 A0A1Q3FQD3 A0A1Q3FQA6 A0A1Q3FQG8 A0A1Q3FQH2 A0A1B6LM42 F4WP32 K7IP31 A0A1B6M0Z3 D4AHY3 N6TVH8 A0A0N0BDQ1
A0A2W1BVC4 A0A194PK39 A0A0L7LSC8 A0A2S2QIG2 J9JV25 A0A1L8DIL8 A0A1L8DIL5 A0A1L8DIJ3 A0A1L8DIK8 A0A212FL47 D7EK15 A0A182MM04 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182S579 A0A182VIR9 A0A182TSI6 V5I8R9 A0A1Y9H2R2 A0A182T9M2 A8CWB1 A0A2S2N8Q6 T1DE67 U5ESS0 A0A2H8TDY2 A0A1B0D4P3 A0A182HJZ0 A0A2M3ZFA3 A0A1Y0AWQ5 W5J543 A0A2M4A0H9 A0A2M4A0I2 E0W0A9 A0A182YLM9 A0A182QT48 A0A182VS58 T1DG93 A0A1Y1LWD0 A0A084VDW1 J3JXM2 N6SYK9 A0A2M4A1M5 A0A2M4BMJ9 A0A2M3ZF95 A0A182FKZ4 A0A1W4XAC6 A0A1L8DF28 U4UIC9 A0A1L8DF43 A0A139WCV1 A0A088ABY6 D7EK13 A0A2M3ZFG5 A0A034W685 A0A0J7L1I9 A0A1W4W7Z0 A0A1B0CZM5 A0A1J1IWB4 A0A1B6IS32 A0A195CMF0 A0A0C9RE51 E9IQR3 A0A1W4XJX1 A0A2A3E9H1 A0A336M0J9 A0A0J7NVW1 A0A0T6BH34 Q16SU5 W8BDP5 A0A0A1WDI7 B0WZW7 A0A0A1WV08 D4AHX1 W8B3Q4 A0A1B6L0T0 A0A1B6MT94 A0A182J8U1 A0A1B6M1S5 A0A2P8Z217 A0A1B6IVC7 A0A2P8Z215 A0A0K8W3N6 A0A1Q3FQD3 A0A1Q3FQA6 A0A1Q3FQG8 A0A1Q3FQH2 A0A1B6LM42 F4WP32 K7IP31 A0A1B6M0Z3 D4AHY3 N6TVH8 A0A0N0BDQ1
Pubmed
EMBL
BABH01016691
BABH01016692
BABH01016693
BABH01016694
BABH01016697
BABH01016696
+ More
ODYU01003112 SOQ41455.1 NWSH01000916 PCG73551.1 PCG73552.1 KZ149936 PZC77147.1 KQ459601 KPI93687.1 JTDY01000189 KOB78385.1 GGMS01007769 MBY76972.1 ABLF02036056 ABLF02036059 GFDF01007791 JAV06293.1 GFDF01007792 JAV06292.1 GFDF01007785 JAV06299.1 GFDF01007786 JAV06298.1 AGBW02007856 OWR54475.1 DS497725 EFA12961.1 AXCM01000233 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 GALX01004205 JAB64261.1 EU124608 ABV60326.1 GGMR01000900 MBY13519.1 GALA01001167 JAA93685.1 GANO01003127 JAB56744.1 GFXV01000127 MBW11932.1 AJVK01011560 APCN01002224 GGFM01006414 MBW27165.1 KY921827 ART29416.1 ADMH02002130 ETN58528.1 GGFK01000993 MBW34314.1 GGFK01000934 MBW34255.1 DS235858 EEB19065.1 AXCN02000090 GAMD01002846 JAA98744.1 GEZM01048061 JAV76640.1 ATLV01011809 KE524723 KFB36155.1 BT127991 AEE62953.1 APGK01058788 APGK01058789 KB741292 ENN70298.1 GGFK01001330 MBW34651.1 GGFJ01004857 MBW53998.1 GGFM01006410 MBW27161.1 GFDF01009038 JAV05046.1 KB632334 ERL92772.1 GFDF01009023 JAV05061.1 KQ971364 KYB25661.1 EFA12963.1 GGFM01006409 MBW27160.1 GAKP01008753 JAC50199.1 LBMM01001313 KMQ96531.1 AJVK01020886 CVRI01000063 CRL04571.1 GECU01017961 JAS89745.1 KQ977574 KYN01893.1 GBYB01014839 JAG84606.1 GL764900 EFZ17069.1 KZ288338 PBC27839.1 UFQT01000389 SSX23856.1 KMQ96530.1 LJIG01000288 KRT86652.1 CH477665 EAT37543.1 GAMC01018656 JAB87899.1 GBXI01017546 JAC96745.1 DS232219 EDS37792.1 GBXI01011595 GBXI01005185 JAD02697.1 JAD09107.1 AB549999 BAI83420.1 GAMC01018654 JAB87901.1 GEBQ01022823 JAT17154.1 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 GEBQ01010102 JAT29875.1 PYGN01000235 PSN50541.1 GECU01016809 JAS90897.1 PSN50537.1 GDHF01014137 GDHF01006620 JAI38177.1 JAI45694.1 GFDL01005268 JAV29777.1 GFDL01005255 JAV29790.1 GFDL01005309 JAV29736.1 GFDL01005283 JAV29762.1 GEBQ01015167 JAT24810.1 GL888243 EGI63963.1 GEBQ01010386 JAT29591.1 AB550011 BAI83432.1 APGK01058785 KI209303 ENN70297.1 ERL96014.1 KQ435851 KOX70761.1
ODYU01003112 SOQ41455.1 NWSH01000916 PCG73551.1 PCG73552.1 KZ149936 PZC77147.1 KQ459601 KPI93687.1 JTDY01000189 KOB78385.1 GGMS01007769 MBY76972.1 ABLF02036056 ABLF02036059 GFDF01007791 JAV06293.1 GFDF01007792 JAV06292.1 GFDF01007785 JAV06299.1 GFDF01007786 JAV06298.1 AGBW02007856 OWR54475.1 DS497725 EFA12961.1 AXCM01000233 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 GALX01004205 JAB64261.1 EU124608 ABV60326.1 GGMR01000900 MBY13519.1 GALA01001167 JAA93685.1 GANO01003127 JAB56744.1 GFXV01000127 MBW11932.1 AJVK01011560 APCN01002224 GGFM01006414 MBW27165.1 KY921827 ART29416.1 ADMH02002130 ETN58528.1 GGFK01000993 MBW34314.1 GGFK01000934 MBW34255.1 DS235858 EEB19065.1 AXCN02000090 GAMD01002846 JAA98744.1 GEZM01048061 JAV76640.1 ATLV01011809 KE524723 KFB36155.1 BT127991 AEE62953.1 APGK01058788 APGK01058789 KB741292 ENN70298.1 GGFK01001330 MBW34651.1 GGFJ01004857 MBW53998.1 GGFM01006410 MBW27161.1 GFDF01009038 JAV05046.1 KB632334 ERL92772.1 GFDF01009023 JAV05061.1 KQ971364 KYB25661.1 EFA12963.1 GGFM01006409 MBW27160.1 GAKP01008753 JAC50199.1 LBMM01001313 KMQ96531.1 AJVK01020886 CVRI01000063 CRL04571.1 GECU01017961 JAS89745.1 KQ977574 KYN01893.1 GBYB01014839 JAG84606.1 GL764900 EFZ17069.1 KZ288338 PBC27839.1 UFQT01000389 SSX23856.1 KMQ96530.1 LJIG01000288 KRT86652.1 CH477665 EAT37543.1 GAMC01018656 JAB87899.1 GBXI01017546 JAC96745.1 DS232219 EDS37792.1 GBXI01011595 GBXI01005185 JAD02697.1 JAD09107.1 AB549999 BAI83420.1 GAMC01018654 JAB87901.1 GEBQ01022823 JAT17154.1 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 GEBQ01010102 JAT29875.1 PYGN01000235 PSN50541.1 GECU01016809 JAS90897.1 PSN50537.1 GDHF01014137 GDHF01006620 JAI38177.1 JAI45694.1 GFDL01005268 JAV29777.1 GFDL01005255 JAV29790.1 GFDL01005309 JAV29736.1 GFDL01005283 JAV29762.1 GEBQ01015167 JAT24810.1 GL888243 EGI63963.1 GEBQ01010386 JAT29591.1 AB550011 BAI83432.1 APGK01058785 KI209303 ENN70297.1 ERL96014.1 KQ435851 KOX70761.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000007819
UP000007151
+ More
UP000007266 UP000075883 UP000075882 UP000007062 UP000076407 UP000075900 UP000075903 UP000075902 UP000075884 UP000075901 UP000092462 UP000075840 UP000000673 UP000009046 UP000076408 UP000075886 UP000075920 UP000030765 UP000019118 UP000069272 UP000192223 UP000030742 UP000005203 UP000036403 UP000183832 UP000078542 UP000242457 UP000008820 UP000002320 UP000075880 UP000245037 UP000007755 UP000002358 UP000053105
UP000007266 UP000075883 UP000075882 UP000007062 UP000076407 UP000075900 UP000075903 UP000075902 UP000075884 UP000075901 UP000092462 UP000075840 UP000000673 UP000009046 UP000076408 UP000075886 UP000075920 UP000030765 UP000019118 UP000069272 UP000192223 UP000030742 UP000005203 UP000036403 UP000183832 UP000078542 UP000242457 UP000008820 UP000002320 UP000075880 UP000245037 UP000007755 UP000002358 UP000053105
Interpro
CDD
ProteinModelPortal
H9JMJ0
H9JMI8
H9JMI9
A0A2H1VKV6
A0A2A4JNK2
A0A2A4JQ62
+ More
A0A2W1BVC4 A0A194PK39 A0A0L7LSC8 A0A2S2QIG2 J9JV25 A0A1L8DIL8 A0A1L8DIL5 A0A1L8DIJ3 A0A1L8DIK8 A0A212FL47 D7EK15 A0A182MM04 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182S579 A0A182VIR9 A0A182TSI6 V5I8R9 A0A1Y9H2R2 A0A182T9M2 A8CWB1 A0A2S2N8Q6 T1DE67 U5ESS0 A0A2H8TDY2 A0A1B0D4P3 A0A182HJZ0 A0A2M3ZFA3 A0A1Y0AWQ5 W5J543 A0A2M4A0H9 A0A2M4A0I2 E0W0A9 A0A182YLM9 A0A182QT48 A0A182VS58 T1DG93 A0A1Y1LWD0 A0A084VDW1 J3JXM2 N6SYK9 A0A2M4A1M5 A0A2M4BMJ9 A0A2M3ZF95 A0A182FKZ4 A0A1W4XAC6 A0A1L8DF28 U4UIC9 A0A1L8DF43 A0A139WCV1 A0A088ABY6 D7EK13 A0A2M3ZFG5 A0A034W685 A0A0J7L1I9 A0A1W4W7Z0 A0A1B0CZM5 A0A1J1IWB4 A0A1B6IS32 A0A195CMF0 A0A0C9RE51 E9IQR3 A0A1W4XJX1 A0A2A3E9H1 A0A336M0J9 A0A0J7NVW1 A0A0T6BH34 Q16SU5 W8BDP5 A0A0A1WDI7 B0WZW7 A0A0A1WV08 D4AHX1 W8B3Q4 A0A1B6L0T0 A0A1B6MT94 A0A182J8U1 A0A1B6M1S5 A0A2P8Z217 A0A1B6IVC7 A0A2P8Z215 A0A0K8W3N6 A0A1Q3FQD3 A0A1Q3FQA6 A0A1Q3FQG8 A0A1Q3FQH2 A0A1B6LM42 F4WP32 K7IP31 A0A1B6M0Z3 D4AHY3 N6TVH8 A0A0N0BDQ1
A0A2W1BVC4 A0A194PK39 A0A0L7LSC8 A0A2S2QIG2 J9JV25 A0A1L8DIL8 A0A1L8DIL5 A0A1L8DIJ3 A0A1L8DIK8 A0A212FL47 D7EK15 A0A182MM04 A0A182KX77 A0NBZ0 A0A1Y9J057 A0A182S579 A0A182VIR9 A0A182TSI6 V5I8R9 A0A1Y9H2R2 A0A182T9M2 A8CWB1 A0A2S2N8Q6 T1DE67 U5ESS0 A0A2H8TDY2 A0A1B0D4P3 A0A182HJZ0 A0A2M3ZFA3 A0A1Y0AWQ5 W5J543 A0A2M4A0H9 A0A2M4A0I2 E0W0A9 A0A182YLM9 A0A182QT48 A0A182VS58 T1DG93 A0A1Y1LWD0 A0A084VDW1 J3JXM2 N6SYK9 A0A2M4A1M5 A0A2M4BMJ9 A0A2M3ZF95 A0A182FKZ4 A0A1W4XAC6 A0A1L8DF28 U4UIC9 A0A1L8DF43 A0A139WCV1 A0A088ABY6 D7EK13 A0A2M3ZFG5 A0A034W685 A0A0J7L1I9 A0A1W4W7Z0 A0A1B0CZM5 A0A1J1IWB4 A0A1B6IS32 A0A195CMF0 A0A0C9RE51 E9IQR3 A0A1W4XJX1 A0A2A3E9H1 A0A336M0J9 A0A0J7NVW1 A0A0T6BH34 Q16SU5 W8BDP5 A0A0A1WDI7 B0WZW7 A0A0A1WV08 D4AHX1 W8B3Q4 A0A1B6L0T0 A0A1B6MT94 A0A182J8U1 A0A1B6M1S5 A0A2P8Z217 A0A1B6IVC7 A0A2P8Z215 A0A0K8W3N6 A0A1Q3FQD3 A0A1Q3FQA6 A0A1Q3FQG8 A0A1Q3FQH2 A0A1B6LM42 F4WP32 K7IP31 A0A1B6M0Z3 D4AHY3 N6TVH8 A0A0N0BDQ1
PDB
5EQI
E-value=3.60408e-12,
Score=166
Ontologies
GO
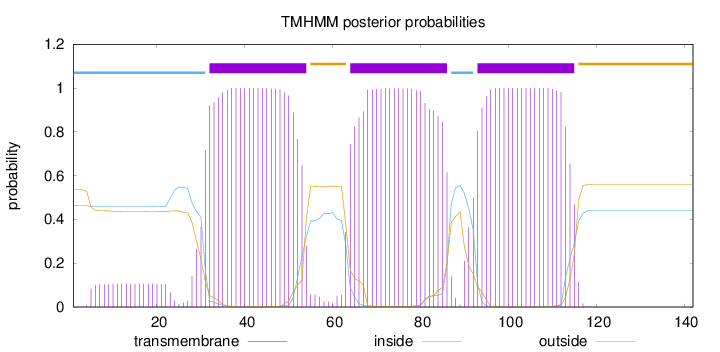
Topology
Length:
142
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
69.78138
Exp number, first 60 AAs:
25.03381
Total prob of N-in:
0.46341
POSSIBLE N-term signal
sequence
inside
1 - 31
TMhelix
32 - 54
outside
55 - 63
TMhelix
64 - 86
inside
87 - 92
TMhelix
93 - 115
outside
116 - 142
Population Genetic Test Statistics
Pi
143.202915
Theta
171.397433
Tajima's D
0.600772
CLR
0.523862
CSRT
0.541372931353432
Interpretation
Uncertain
