Pre Gene Modal
BGIBMGA010801
Annotation
Y+L_amino_acid_transporter_2_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.985
Sequence
CDS
ATGGAGGGCGACAACGTCAGCAAAGACGGGGGGGCTCTGGAAGCCAAGATGTCCTTGCTGAACGGGTGCACGGTTATCGTCGGCTCCATCATCGGAAGTGGAATCTTCGTATCGCCAACTGGAGTCCTCAAGTATACAGGTTCCGTGAACGCTAGTCTTCTTGTTTGGATTGCCTCAGGAGTTTTTAGTATGATCGGCGCCTACTGCTATGCTGAACTTGGGACGATGATCAGGCTAAGCGGCGCGGACTACGCTTACATCATGGAGACTTTTGGACCGTTCGCTGCTTTCGTGAGGCTCTGGATCGAGTGCATGATTGTACGGCCTTGTTCGCAGGCCATAGTTGCGTTAACATTCAGCGTGTACGTGTTGAAGCCTCTGTTTCCGGAGTGCGACCCTCCCGAAGACGCGACCAGATTACTCGCGGCTTGCTGCATATTGTTGCTGACCTTCGTGAATTGCTGGTCGGTACGCGCAGCTACTTTGGTACAGGACTGGTTCACGTACGCCAAGCTCCTGGCACTTTTCATCATCATCGCAGCCGGGATCTATCAACTGTGCAGAGGTGAGAACTACTTGAACTTTATAATAGAGGAGTTGAAGGATCCCGTCCGCAACCTGCCCCGCGCCATCGCCATCTCGTGCGCGCTGGTCACCATCGTCTACACGCTCACCAACGTCGCCTTCTACACCACGCTCTCGCCCACCGAGGTGCTGGGCTCAGCAGCGGTAGCAGTAACGTTTGCCGAGCGCTTGTTCGGTTGGTTCGCCCTGTCCATTCCGTTGTTCGTAGCCGCCTCCACTTTCGGAGCTGTTAATGGTGTTCTGCTGACTTCTTCTAGGTTGTTCTACGCGGGCGCCGCGCAGGGCCAGATGCCGGCCATGTTGACGATGGTGACGTCACGCGCCACGCCCGCGCCCGCCGTGCTGGCCGTGGCGCTGCTGTCGCTGCTCTACCTCACCGTCTCCGACATATACGCGCTCATCAACTACGTCGGATTCGCTACCTGGTTAAGTATTGGAGCCGCGGTACTCTGTCTGCCTGTGCTGAGATACACGAAGCCAGATCTCGATCGACCCATTAAAGTTAATCTGTTCTTCCCGGTGATCTACATCATCTGCACGATCCTGGTGGTGGCTGTACCGGCGGTGGCGTCACCGGCCGAGACCGGCATCGGCTGCCTCATGATATTCACCGCTGCGCCGGTCTACCTGCTACTCTTGCACCCCAGGACTAGAATTAACTTCCTCGCTAATATTATGCACGGCGCAACCTACTTGGTACAAAAGCTAACGCTCTCCGTTAGACCGAAAACGAAGTGA
Protein
MEGDNVSKDGGALEAKMSLLNGCTVIVGSIIGSGIFVSPTGVLKYTGSVNASLLVWIASGVFSMIGAYCYAELGTMIRLSGADYAYIMETFGPFAAFVRLWIECMIVRPCSQAIVALTFSVYVLKPLFPECDPPEDATRLLAACCILLLTFVNCWSVRAATLVQDWFTYAKLLALFIIIAAGIYQLCRGENYLNFIIEELKDPVRNLPRAIAISCALVTIVYTLTNVAFYTTLSPTEVLGSAAVAVTFAERLFGWFALSIPLFVAASTFGAVNGVLLTSSRLFYAGAAQGQMPAMLTMVTSRATPAPAVLAVALLSLLYLTVSDIYALINYVGFATWLSIGAAVLCLPVLRYTKPDLDRPIKVNLFFPVIYIICTILVVAVPAVASPAETGIGCLMIFTAAPVYLLLLHPRTRINFLANIMHGATYLVQKLTLSVRPKTK
Summary
Uniprot
H9JMP8
A0A2W1BWH3
A0A2H1VZD6
A0A3S2P4K4
A0A194PL09
A0A212F8K2
+ More
A0A2A4JND8 A0A0N1IQ90 A0A1B6E8G3 A0A154NXY2 A0A0M8ZYK9 A0A146L8A0 E2BNK6 A0A2M4AUL6 A0A182YB76 A0A084VWJ6 A0A182WSI9 Q7PXW3 A0A182HLE1 A0A182V0Y8 A0A023F5K5 W8C7D3 A0A2M4BMS8 A0A069DUF2 A0A0K8SP07 A0A224XP93 A0A2J7QHY0 A0A182ISU0 Q17HW5 A0A034VB41 A0A0K8W3S2 V9IFX2 A0A0V0G4I6 A0A182GYB6 A0A1Q3FUK0 A0A1Q3FUH7 A0A1Y1KEC8 A0A2M4AU82 A0A182VZD1 A0A182R611 A0A182LRK3 W5J8I0 A0A1J1HVT3 K7IMA0 A0A182FQ35 A0A182Q1T0 A0A182TND8 D6WZJ6 A0A182L9S7 B4PND5 A0A1W4WD86 A0A310SJQ3 A0A2J7QHX1 A0A0R1E6D0 A0A067QZP3 A0A1Q3FTY9 A0A1Q3FU07 A0A182S880 A0A0P5RWF1 A0A0P5HAI8 A0A1A9V7M7 A0A0P6C1G4 E9HBN1 A0A0N8DDN0 A0A0A9X4J3 A0A0P5RD75 A0A0P5ZCX4 A0A0N7ZYD1 A0A0P5VE32 A0A0P4Z4M8 A0A2P2I2B2 A0A0P4WLV6 A0A1D2N3T2 A0A0P6I0U7 A0A0P5YLW4 A0A1E1XG15 A0A2R5L9N8 A0A336MMR8 A0A0P5P377 A0A2C9K9L6 A0A1S3IH31 K1RCM5 A0A210PSU8 A0A0P5WZM5 A0A2C9K9T1 S9X9A7 A0A1W4WE76 A0A085N859 A0A1I7SR35 A0A077ZE72 A0A2J8VVA0 H3DK93 A0A0K0D2I8 A0A3B3SSR4 A0A3N0YI50 A0A3B4E2K3 A0A088ADE9 F1R8V8 A0A3Q1BIG0 W5KUL3
A0A2A4JND8 A0A0N1IQ90 A0A1B6E8G3 A0A154NXY2 A0A0M8ZYK9 A0A146L8A0 E2BNK6 A0A2M4AUL6 A0A182YB76 A0A084VWJ6 A0A182WSI9 Q7PXW3 A0A182HLE1 A0A182V0Y8 A0A023F5K5 W8C7D3 A0A2M4BMS8 A0A069DUF2 A0A0K8SP07 A0A224XP93 A0A2J7QHY0 A0A182ISU0 Q17HW5 A0A034VB41 A0A0K8W3S2 V9IFX2 A0A0V0G4I6 A0A182GYB6 A0A1Q3FUK0 A0A1Q3FUH7 A0A1Y1KEC8 A0A2M4AU82 A0A182VZD1 A0A182R611 A0A182LRK3 W5J8I0 A0A1J1HVT3 K7IMA0 A0A182FQ35 A0A182Q1T0 A0A182TND8 D6WZJ6 A0A182L9S7 B4PND5 A0A1W4WD86 A0A310SJQ3 A0A2J7QHX1 A0A0R1E6D0 A0A067QZP3 A0A1Q3FTY9 A0A1Q3FU07 A0A182S880 A0A0P5RWF1 A0A0P5HAI8 A0A1A9V7M7 A0A0P6C1G4 E9HBN1 A0A0N8DDN0 A0A0A9X4J3 A0A0P5RD75 A0A0P5ZCX4 A0A0N7ZYD1 A0A0P5VE32 A0A0P4Z4M8 A0A2P2I2B2 A0A0P4WLV6 A0A1D2N3T2 A0A0P6I0U7 A0A0P5YLW4 A0A1E1XG15 A0A2R5L9N8 A0A336MMR8 A0A0P5P377 A0A2C9K9L6 A0A1S3IH31 K1RCM5 A0A210PSU8 A0A0P5WZM5 A0A2C9K9T1 S9X9A7 A0A1W4WE76 A0A085N859 A0A1I7SR35 A0A077ZE72 A0A2J8VVA0 H3DK93 A0A0K0D2I8 A0A3B3SSR4 A0A3N0YI50 A0A3B4E2K3 A0A088ADE9 F1R8V8 A0A3Q1BIG0 W5KUL3
Pubmed
19121390
28756777
26354079
22118469
26823975
20798317
+ More
25244985 24438588 12364791 14747013 17210077 25474469 24495485 26334808 17510324 25348373 26483478 28004739 20920257 23761445 20075255 18362917 19820115 20966253 17994087 17550304 24845553 21292972 25401762 27289101 28503490 15562597 22992520 28812685 23149746 24929829 21909270 15496914 29240929 23594743 25329095
25244985 24438588 12364791 14747013 17210077 25474469 24495485 26334808 17510324 25348373 26483478 28004739 20920257 23761445 20075255 18362917 19820115 20966253 17994087 17550304 24845553 21292972 25401762 27289101 28503490 15562597 22992520 28812685 23149746 24929829 21909270 15496914 29240929 23594743 25329095
EMBL
BABH01016685
KZ149936
PZC77150.1
ODYU01005399
SOQ46191.1
RSAL01000483
+ More
RVE41571.1 KQ459601 KPI93683.1 AGBW02009706 OWR50075.1 NWSH01000916 PCG73547.1 KQ459910 KPJ19339.1 GEDC01003154 JAS34144.1 KQ434766 KZC03740.1 KQ435812 KOX72726.1 GDHC01014814 JAQ03815.1 GL449414 EFN82785.1 GGFK01011091 MBW44412.1 ATLV01017623 KE525179 KFB42340.1 AAAB01008987 EAA00996.3 APCN01000894 GBBI01002273 JAC16439.1 GAMC01008092 GAMC01008091 JAB98464.1 GGFJ01005100 MBW54241.1 GBGD01001304 JAC87585.1 GBRD01010955 JAG54869.1 GFTR01006607 JAW09819.1 NEVH01013964 PNF28194.1 CH477245 EAT46224.1 GAKP01019248 JAC39704.1 GDHF01016415 GDHF01006573 JAI35899.1 JAI45741.1 JR045423 AEY59975.1 GECL01003524 JAP02600.1 JXUM01021304 KQ560596 KXJ81706.1 GFDL01003827 JAV31218.1 GFDL01003828 JAV31217.1 GEZM01087142 JAV58888.1 GGFK01010827 MBW44148.1 AXCM01001257 ADMH02002100 ETN59180.1 CVRI01000023 CRK92091.1 AXCN02000266 KQ971372 EFA09694.1 CM000160 EDW99217.2 KQ760549 OAD59898.1 PNF28195.1 KRK04731.1 KRK04732.1 KK852854 KDR14996.1 GFDL01003964 JAV31081.1 GFDL01003965 JAV31080.1 GDIP01199123 GDIQ01095345 GDIQ01081958 LRGB01000084 JAJ24279.1 JAL56381.1 KZS21113.1 GDIQ01235754 GDIP01050560 JAK15971.1 JAM53155.1 GDIP01006122 JAM97593.1 GL732616 EFX70890.1 GDIP01044007 JAM59708.1 GBHO01043162 GBHO01043161 GBHO01043160 GBHO01030822 GBHO01029886 GBHO01001093 GBHO01001092 GBHO01001090 GBHO01001089 JAG00442.1 JAG00443.1 JAG00444.1 JAG12782.1 JAG13718.1 JAG42511.1 JAG42512.1 JAG42514.1 JAG42515.1 GDIQ01171017 GDIQ01108640 JAL43086.1 GDIP01045517 JAM58198.1 GDIP01200489 GDIQ01051066 JAJ22913.1 JAN43671.1 GDIP01101638 JAM02077.1 GDIP01219486 JAJ03916.1 IACF01002478 LAB68131.1 GDRN01062307 JAI65093.1 LJIJ01000246 ODM99929.1 GDIQ01011178 JAN83559.1 GDIP01055956 JAM47759.1 GFAC01000983 JAT98205.1 GGLE01002086 MBY06212.1 UFQS01001767 UFQT01001767 SSX12273.1 SSX31724.1 GDIQ01138638 JAL13088.1 JH816265 EKC38980.1 NEDP02005521 OWF39558.1 GDIP01091456 JAM12259.1 KB016774 EPY84179.1 KL363219 KL367534 KFD53159.1 KFD65655.1 HG806385 CDW58641.1 NDHI03003408 PNJ61451.1 RJVU01042551 ROL45551.1 CR352328
RVE41571.1 KQ459601 KPI93683.1 AGBW02009706 OWR50075.1 NWSH01000916 PCG73547.1 KQ459910 KPJ19339.1 GEDC01003154 JAS34144.1 KQ434766 KZC03740.1 KQ435812 KOX72726.1 GDHC01014814 JAQ03815.1 GL449414 EFN82785.1 GGFK01011091 MBW44412.1 ATLV01017623 KE525179 KFB42340.1 AAAB01008987 EAA00996.3 APCN01000894 GBBI01002273 JAC16439.1 GAMC01008092 GAMC01008091 JAB98464.1 GGFJ01005100 MBW54241.1 GBGD01001304 JAC87585.1 GBRD01010955 JAG54869.1 GFTR01006607 JAW09819.1 NEVH01013964 PNF28194.1 CH477245 EAT46224.1 GAKP01019248 JAC39704.1 GDHF01016415 GDHF01006573 JAI35899.1 JAI45741.1 JR045423 AEY59975.1 GECL01003524 JAP02600.1 JXUM01021304 KQ560596 KXJ81706.1 GFDL01003827 JAV31218.1 GFDL01003828 JAV31217.1 GEZM01087142 JAV58888.1 GGFK01010827 MBW44148.1 AXCM01001257 ADMH02002100 ETN59180.1 CVRI01000023 CRK92091.1 AXCN02000266 KQ971372 EFA09694.1 CM000160 EDW99217.2 KQ760549 OAD59898.1 PNF28195.1 KRK04731.1 KRK04732.1 KK852854 KDR14996.1 GFDL01003964 JAV31081.1 GFDL01003965 JAV31080.1 GDIP01199123 GDIQ01095345 GDIQ01081958 LRGB01000084 JAJ24279.1 JAL56381.1 KZS21113.1 GDIQ01235754 GDIP01050560 JAK15971.1 JAM53155.1 GDIP01006122 JAM97593.1 GL732616 EFX70890.1 GDIP01044007 JAM59708.1 GBHO01043162 GBHO01043161 GBHO01043160 GBHO01030822 GBHO01029886 GBHO01001093 GBHO01001092 GBHO01001090 GBHO01001089 JAG00442.1 JAG00443.1 JAG00444.1 JAG12782.1 JAG13718.1 JAG42511.1 JAG42512.1 JAG42514.1 JAG42515.1 GDIQ01171017 GDIQ01108640 JAL43086.1 GDIP01045517 JAM58198.1 GDIP01200489 GDIQ01051066 JAJ22913.1 JAN43671.1 GDIP01101638 JAM02077.1 GDIP01219486 JAJ03916.1 IACF01002478 LAB68131.1 GDRN01062307 JAI65093.1 LJIJ01000246 ODM99929.1 GDIQ01011178 JAN83559.1 GDIP01055956 JAM47759.1 GFAC01000983 JAT98205.1 GGLE01002086 MBY06212.1 UFQS01001767 UFQT01001767 SSX12273.1 SSX31724.1 GDIQ01138638 JAL13088.1 JH816265 EKC38980.1 NEDP02005521 OWF39558.1 GDIP01091456 JAM12259.1 KB016774 EPY84179.1 KL363219 KL367534 KFD53159.1 KFD65655.1 HG806385 CDW58641.1 NDHI03003408 PNJ61451.1 RJVU01042551 ROL45551.1 CR352328
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000218220
UP000053240
+ More
UP000076502 UP000053105 UP000008237 UP000076408 UP000030765 UP000076407 UP000007062 UP000075840 UP000075903 UP000235965 UP000075880 UP000008820 UP000069940 UP000249989 UP000075920 UP000075900 UP000075883 UP000000673 UP000183832 UP000002358 UP000069272 UP000075886 UP000075902 UP000007266 UP000075882 UP000002282 UP000192223 UP000027135 UP000075901 UP000076858 UP000078200 UP000000305 UP000094527 UP000076420 UP000085678 UP000005408 UP000242188 UP000095284 UP000030665 UP000007303 UP000035642 UP000261540 UP000261440 UP000005203 UP000000437 UP000257160 UP000018467
UP000076502 UP000053105 UP000008237 UP000076408 UP000030765 UP000076407 UP000007062 UP000075840 UP000075903 UP000235965 UP000075880 UP000008820 UP000069940 UP000249989 UP000075920 UP000075900 UP000075883 UP000000673 UP000183832 UP000002358 UP000069272 UP000075886 UP000075902 UP000007266 UP000075882 UP000002282 UP000192223 UP000027135 UP000075901 UP000076858 UP000078200 UP000000305 UP000094527 UP000076420 UP000085678 UP000005408 UP000242188 UP000095284 UP000030665 UP000007303 UP000035642 UP000261540 UP000261440 UP000005203 UP000000437 UP000257160 UP000018467
PRIDE
Pfam
PF13520 AA_permease_2
Interpro
IPR002293
AA/rel_permease1
ProteinModelPortal
H9JMP8
A0A2W1BWH3
A0A2H1VZD6
A0A3S2P4K4
A0A194PL09
A0A212F8K2
+ More
A0A2A4JND8 A0A0N1IQ90 A0A1B6E8G3 A0A154NXY2 A0A0M8ZYK9 A0A146L8A0 E2BNK6 A0A2M4AUL6 A0A182YB76 A0A084VWJ6 A0A182WSI9 Q7PXW3 A0A182HLE1 A0A182V0Y8 A0A023F5K5 W8C7D3 A0A2M4BMS8 A0A069DUF2 A0A0K8SP07 A0A224XP93 A0A2J7QHY0 A0A182ISU0 Q17HW5 A0A034VB41 A0A0K8W3S2 V9IFX2 A0A0V0G4I6 A0A182GYB6 A0A1Q3FUK0 A0A1Q3FUH7 A0A1Y1KEC8 A0A2M4AU82 A0A182VZD1 A0A182R611 A0A182LRK3 W5J8I0 A0A1J1HVT3 K7IMA0 A0A182FQ35 A0A182Q1T0 A0A182TND8 D6WZJ6 A0A182L9S7 B4PND5 A0A1W4WD86 A0A310SJQ3 A0A2J7QHX1 A0A0R1E6D0 A0A067QZP3 A0A1Q3FTY9 A0A1Q3FU07 A0A182S880 A0A0P5RWF1 A0A0P5HAI8 A0A1A9V7M7 A0A0P6C1G4 E9HBN1 A0A0N8DDN0 A0A0A9X4J3 A0A0P5RD75 A0A0P5ZCX4 A0A0N7ZYD1 A0A0P5VE32 A0A0P4Z4M8 A0A2P2I2B2 A0A0P4WLV6 A0A1D2N3T2 A0A0P6I0U7 A0A0P5YLW4 A0A1E1XG15 A0A2R5L9N8 A0A336MMR8 A0A0P5P377 A0A2C9K9L6 A0A1S3IH31 K1RCM5 A0A210PSU8 A0A0P5WZM5 A0A2C9K9T1 S9X9A7 A0A1W4WE76 A0A085N859 A0A1I7SR35 A0A077ZE72 A0A2J8VVA0 H3DK93 A0A0K0D2I8 A0A3B3SSR4 A0A3N0YI50 A0A3B4E2K3 A0A088ADE9 F1R8V8 A0A3Q1BIG0 W5KUL3
A0A2A4JND8 A0A0N1IQ90 A0A1B6E8G3 A0A154NXY2 A0A0M8ZYK9 A0A146L8A0 E2BNK6 A0A2M4AUL6 A0A182YB76 A0A084VWJ6 A0A182WSI9 Q7PXW3 A0A182HLE1 A0A182V0Y8 A0A023F5K5 W8C7D3 A0A2M4BMS8 A0A069DUF2 A0A0K8SP07 A0A224XP93 A0A2J7QHY0 A0A182ISU0 Q17HW5 A0A034VB41 A0A0K8W3S2 V9IFX2 A0A0V0G4I6 A0A182GYB6 A0A1Q3FUK0 A0A1Q3FUH7 A0A1Y1KEC8 A0A2M4AU82 A0A182VZD1 A0A182R611 A0A182LRK3 W5J8I0 A0A1J1HVT3 K7IMA0 A0A182FQ35 A0A182Q1T0 A0A182TND8 D6WZJ6 A0A182L9S7 B4PND5 A0A1W4WD86 A0A310SJQ3 A0A2J7QHX1 A0A0R1E6D0 A0A067QZP3 A0A1Q3FTY9 A0A1Q3FU07 A0A182S880 A0A0P5RWF1 A0A0P5HAI8 A0A1A9V7M7 A0A0P6C1G4 E9HBN1 A0A0N8DDN0 A0A0A9X4J3 A0A0P5RD75 A0A0P5ZCX4 A0A0N7ZYD1 A0A0P5VE32 A0A0P4Z4M8 A0A2P2I2B2 A0A0P4WLV6 A0A1D2N3T2 A0A0P6I0U7 A0A0P5YLW4 A0A1E1XG15 A0A2R5L9N8 A0A336MMR8 A0A0P5P377 A0A2C9K9L6 A0A1S3IH31 K1RCM5 A0A210PSU8 A0A0P5WZM5 A0A2C9K9T1 S9X9A7 A0A1W4WE76 A0A085N859 A0A1I7SR35 A0A077ZE72 A0A2J8VVA0 H3DK93 A0A0K0D2I8 A0A3B3SSR4 A0A3N0YI50 A0A3B4E2K3 A0A088ADE9 F1R8V8 A0A3Q1BIG0 W5KUL3
PDB
6IRT
E-value=9.64914e-83,
Score=782
Ontologies
GO
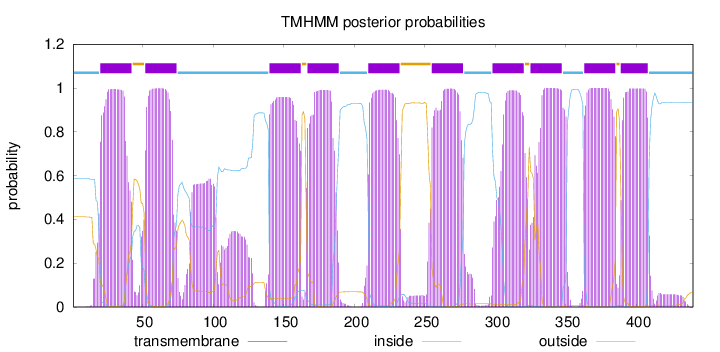
Topology
Length:
440
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
237.46676
Exp number, first 60 AAs:
31.72982
Total prob of N-in:
0.58763
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 51
TMhelix
52 - 74
inside
75 - 139
TMhelix
140 - 162
outside
163 - 166
TMhelix
167 - 189
inside
190 - 209
TMhelix
210 - 232
outside
233 - 254
TMhelix
255 - 277
inside
278 - 297
TMhelix
298 - 320
outside
321 - 324
TMhelix
325 - 347
inside
348 - 362
TMhelix
363 - 385
outside
386 - 388
TMhelix
389 - 408
inside
409 - 440
Population Genetic Test Statistics
Pi
24.891324
Theta
169.173631
Tajima's D
0.558603
CLR
0.403805
CSRT
0.532073396330183
Interpretation
Uncertain
