Gene
KWMTBOMO15524
Pre Gene Modal
BGIBMGA010748
Annotation
PREDICTED:_nicotinamide/nicotinic_acid_mononucleotide_adenylyltransferase_3_[Bombyx_mori]
Full name
Nicotinamide-nucleotide adenylyltransferase
Alternative Name
Nicotinate-nucleotide adenylyltransferase
Location in the cell
Cytoplasmic Reliability : 1.13 Mitochondrial Reliability : 1.595
Sequence
CDS
ATGTTCGAAATTGCCCGTGATTACATACATTCGTTTGGTATTGGTTCAGTAGTCGGCGGAATCGTTTCTCCTGTCCACGACGCTTACGGCAAAAAGGATTTAGTTGCTTCGCATCACAGAATTAGCATGTTGAAATTAGCCTTGCGTTCCTCAAACTGGATCAAAGTATCGGAATGGGAATCTCGTCAGGCTGGCTGGACAAGAACCAGACTGTCCTTAAAATACCATCAAGATATAATAAATGGATACAAATCAAATTTAGATCCAGATTCAGCGGAGTGGTTGCCAGATGACGTTCTCAATGTGAACAGCTATGATGAGACTGACAACTTTCCTCTAATGACGAATGGTAACTATCAAGAACGGGTGACAGTAAAGCTGCTCTGCGGTGCTGATCTTCTGGAGTCCTTCGCCACCCCGGGACTTTGGTCTGATGAAGATATCGAAGCAATAGTCGGTACGCATGGCTTAGTGGTGGTCACCAGGGCAGGCAGTAACCCCTCCAAGTTTATTCACGAATCTGACATACTTTATAAGCACAGGAGGAACATCCTGCTGGTGCCCAACCACGTGGTCAACGAGGTGAGCTCCACCGTGGTGCGGCGGCTGCTGCGGCGAGGCGAGAGCGCGCGCTACCTCACCGACGACGCCGTGCTCGCCTACGCCGCCCGCCACGCGCTCTACGCCGCCGCGCCCTCCCGCACCGACTCGGTCAGTCCGGTCGACGGCGTGAAACCGAAGCTGGCGTACGTGGACAGGGTGCCCCCCGGCTACGTGCCCGGGAAGGCGGTCAAGATCGTTAGTGACGTCGCCCAGCACGCCACGGCCGGCGACAAGGTGAGTCCGCACCCGTGTGGCGGCGACCCCCCCGCGCGGCCCCCCTCGCCCCCCCGCGGCTCCTCGAGGGGGCGGTGCGCGACCGACGTTAAAACCAGTCGATCGGAAGCAAGCAAGCTGTGCGACAAGGCGAGCGCGGGGCAGGGGTTGGGGGGCGCGGTGCGGGGGCCGGGGGTTGGGGCGCGGGGGTCGCGGAGCTGCGAGGACATCGTGCGTCTGCTGCTGACCAAGCACGGCATCCACGTGCTGAGCGACTCGGAGGCCGTCGTGTGA
Protein
MFEIARDYIHSFGIGSVVGGIVSPVHDAYGKKDLVASHHRISMLKLALRSSNWIKVSEWESRQAGWTRTRLSLKYHQDIINGYKSNLDPDSAEWLPDDVLNVNSYDETDNFPLMTNGNYQERVTVKLLCGADLLESFATPGLWSDEDIEAIVGTHGLVVVTRAGSNPSKFIHESDILYKHRRNILLVPNHVVNEVSSTVVRRLLRRGESARYLTDDAVLAYAARHALYAAAPSRTDSVSPVDGVKPKLAYVDRVPPGYVPGKAVKIVSDVAQHATAGDKVSPHPCGGDPPARPPSPPRGSSRGRCATDVKTSRSEASKLCDKASAGQGLGGAVRGPGVGARGSRSCEDIVRLLLTKHGIHVLSDSEAVV
Summary
Catalytic Activity
ATP + H(+) + nicotinate beta-D-ribonucleotide = deamido-NAD(+) + diphosphate
ATP + beta-nicotinamide D-ribonucleotide + H(+) = diphosphate + NAD(+)
ATP + beta-nicotinamide D-ribonucleotide + H(+) = diphosphate + NAD(+)
Similarity
Belongs to the eukaryotic NMN adenylyltransferase family.
Uniprot
EC Number
2.7.7.1
2.7.7.18
2.7.7.18
Pubmed
EMBL
Proteomes
PRIDE
Pfam
PF01467 CTP_transf_like
Gene 3D
ProteinModelPortal
PDB
1GZU
E-value=5.88866e-36,
Score=378
Ontologies
PATHWAY
GO
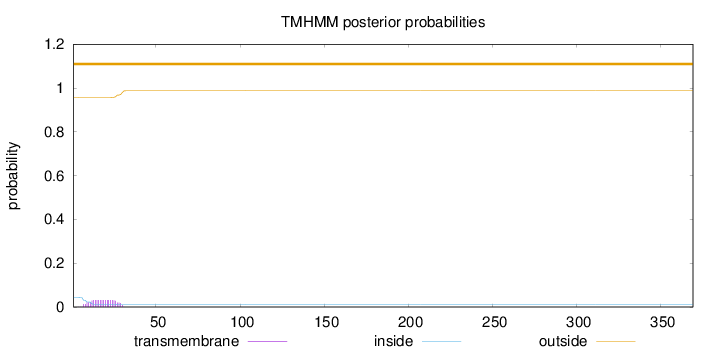
Topology
Length:
369
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.625259999999998
Exp number, first 60 AAs:
0.6245
Total prob of N-in:
0.04320
outside
1 - 369
Population Genetic Test Statistics
Pi
183.238817
Theta
145.756148
Tajima's D
0.468467
CLR
0.457149
CSRT
0.501624918754062
Interpretation
Uncertain
