Gene
KWMTBOMO15522
Pre Gene Modal
BGIBMGA010795
Annotation
PREDICTED:_beta-1?4-galactosyltransferase_7_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.286 PlasmaMembrane Reliability : 1.32
Sequence
CDS
ATGTACATTAGTATTAGCTTGATGGGTTACAGGATAACCAACTCAAGATGTCTCGTTTTGTGCGTAATACTAACTTTTGTGATGGGCTGCTATTTAGCATCTTTGCCCATAGAAGAAGAAAAACCAGCATTGCTTAAGCAATCAGTTGCCGAAATTCCTACCATTAATACAAACAAGAAGCGTCTAGCAATCATCGTTCCGTTCCGGGACCGCTTCGAGGAGCTCCTAGAGTTCGTTCCTCACATGACCGCCTTCCTGAAGAGACAAGAAATTCCATTCCACATCTTTGTGGTTCAACAGAAGGACAGCAACCGTTTCAACAGAGCTTCATTGATAAATGTTGGATTCCTGCAGACTCGTAACAATTTCGAATACATGGCGATGCACGACGTTGATCTTCTACCTTTGAACGACAACCTAAAGTATGAATATCCTGAAGCAGGACCATATCATATCTCTTCACCAGACACCCATCCAAGATATTACTACAAAACGTTCATTGGTGGCATACTGTTAATAAAGAGAGAGCATTATGAGCTGGTCAACGGAATGTCCAATAACTACTGGGGTTGGGGGCTGGAAGACGACGAGTTCTACGTGAGATTGAAGGATGCAGGCCTCACAGTTACGAGGCCAAGCAACATTACTACGGGACGAGAGAACACCTTTAGGCACATGCACGATAAGACTTATCGCAAACGCGACATGCGCAAGTGCTACAACCAGCGCGAGGTGACGCGGCGTCGTGACCGCCGCACCGGACTCCATGACGTGGCCCACCGCGTGCTGAGCCTCCACACCCTCACCATCGACGGGCTGCCGCTGACCGTCCTCAACGTGGAGCTCATCTGCGACCGGAACGCGACCCCGTGGTGTCAGTGCCCAGAACCACCTAAACCTAAAAAGACTTGA
Protein
MYISISLMGYRITNSRCLVLCVILTFVMGCYLASLPIEEEKPALLKQSVAEIPTINTNKKRLAIIVPFRDRFEELLEFVPHMTAFLKRQEIPFHIFVVQQKDSNRFNRASLINVGFLQTRNNFEYMAMHDVDLLPLNDNLKYEYPEAGPYHISSPDTHPRYYYKTFIGGILLIKREHYELVNGMSNNYWGWGLEDDEFYVRLKDAGLTVTRPSNITTGRENTFRHMHDKTYRKRDMRKCYNQREVTRRRDRRTGLHDVAHRVLSLHTLTIDGLPLTVLNVELICDRNATPWCQCPEPPKPKKT
Summary
Uniprot
H9JMP2
A0A2A4IVI1
A0A3S2LU13
A0A2H1WA90
A0A1E1W143
A0A212EZ58
+ More
H9JMP3 A0A0N1ICK8 A0A2A4IVC0 A0A2W1BFI1 E2BNK8 A0A2J7PI40 A0A067QVV9 A0A182L5R8 A0A2M4AJQ8 A0A310SK71 A0A151IKZ1 A0A182TTQ5 A0A182J539 A0A195FHX2 A0A151IVK2 A0A182M291 A0A026WE58 F4X376 A0A151WEJ6 Q170H3 A0A0L7LEQ1 B0X6J8 A0A1J1HPQ6 A0A195BEQ9 A0A182NE87 A0A158NLH9 A0A2M4DL65 A0A1S4FHV9 A0A2M4BVR8 U5ER90 A0A1B6JX31 A0A2A3EB24 A0A232F317 A0A088AQ60 K7J8T5 A0A1Q3FFM7 A0A1W7R8I2 A0A1B6MBL0 A0A182GN55 A0A336M1M5 A0A1B6CJ92 A0A2M3ZC79 A0A1Y1NFZ7 A0A0V0GD08 A0A069DYJ9 A0A0L0C6M0 A0A1B0CMR6 A0A023F434 A0A182K169 A0A182PB48 B4PUZ9 D6X0P7 A0A1I8NF56 B4LX20 B3LWZ2 Q2HPN8 Q2HPP0 B4K7U3 A0A1W4VL56 Q2HPP1 W8BV94 Q2HPP3 A0A034WEA3 A0A3R7MXU4 A0A0K8U4J1 B4NH65 B4K0Z5 A0A0A1WVS4 A0A3B0JXX2 A0A0P4VSM5 E0VV01 A0A1I8NP27 A0A0M4EQ88 A0A0P4VXD1 T1HLB7 A0A182F4E9 Q299T4 A0A0A9Z975 A0A182T624 T1J0Y7 A0A0K8SVC7 A0A1B0BRG9 B4G5Q0 A0A1A9YK92 A0A1A9UN89 Q2HPN9 Q9VBZ9 T1KB34 Q8I6J0 A0A1B0AAB3 N6U3Z6 Q2HPP2 D1H0P4
H9JMP3 A0A0N1ICK8 A0A2A4IVC0 A0A2W1BFI1 E2BNK8 A0A2J7PI40 A0A067QVV9 A0A182L5R8 A0A2M4AJQ8 A0A310SK71 A0A151IKZ1 A0A182TTQ5 A0A182J539 A0A195FHX2 A0A151IVK2 A0A182M291 A0A026WE58 F4X376 A0A151WEJ6 Q170H3 A0A0L7LEQ1 B0X6J8 A0A1J1HPQ6 A0A195BEQ9 A0A182NE87 A0A158NLH9 A0A2M4DL65 A0A1S4FHV9 A0A2M4BVR8 U5ER90 A0A1B6JX31 A0A2A3EB24 A0A232F317 A0A088AQ60 K7J8T5 A0A1Q3FFM7 A0A1W7R8I2 A0A1B6MBL0 A0A182GN55 A0A336M1M5 A0A1B6CJ92 A0A2M3ZC79 A0A1Y1NFZ7 A0A0V0GD08 A0A069DYJ9 A0A0L0C6M0 A0A1B0CMR6 A0A023F434 A0A182K169 A0A182PB48 B4PUZ9 D6X0P7 A0A1I8NF56 B4LX20 B3LWZ2 Q2HPN8 Q2HPP0 B4K7U3 A0A1W4VL56 Q2HPP1 W8BV94 Q2HPP3 A0A034WEA3 A0A3R7MXU4 A0A0K8U4J1 B4NH65 B4K0Z5 A0A0A1WVS4 A0A3B0JXX2 A0A0P4VSM5 E0VV01 A0A1I8NP27 A0A0M4EQ88 A0A0P4VXD1 T1HLB7 A0A182F4E9 Q299T4 A0A0A9Z975 A0A182T624 T1J0Y7 A0A0K8SVC7 A0A1B0BRG9 B4G5Q0 A0A1A9YK92 A0A1A9UN89 Q2HPN9 Q9VBZ9 T1KB34 Q8I6J0 A0A1B0AAB3 N6U3Z6 Q2HPP2 D1H0P4
Pubmed
19121390
22118469
26354079
28756777
20798317
24845553
+ More
20966253 24508170 30249741 21719571 17510324 26227816 21347285 28648823 20075255 26483478 28004739 26334808 26108605 25474469 17994087 17550304 18362917 19820115 25315136 24495485 25348373 25830018 20566863 15632085 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20236943 24052259 26109357 26109356 12215432 23537049 18057021
20966253 24508170 30249741 21719571 17510324 26227816 21347285 28648823 20075255 26483478 28004739 26334808 26108605 25474469 17994087 17550304 18362917 19820115 25315136 24495485 25348373 25830018 20566863 15632085 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20236943 24052259 26109357 26109356 12215432 23537049 18057021
EMBL
BABH01016662
NWSH01007018
PCG63133.1
RSAL01000010
RVE53758.1
ODYU01007305
+ More
SOQ49970.1 GDQN01010334 JAT80720.1 AGBW02011361 OWR46785.1 BABH01016664 KQ460952 KPJ10365.1 PCG63132.1 KZ150188 PZC72434.1 GL449414 EFN82787.1 NEVH01025129 PNF15988.1 KK852895 KDR14189.1 GGFK01007651 MBW40972.1 KQ760478 OAD60206.1 KQ977151 KYN05280.1 KQ981606 KYN39589.1 KQ980895 KYN11764.1 AXCM01002140 KK107250 QOIP01000002 EZA54390.1 RLU25841.1 GL888609 EGI59057.1 KQ983238 KYQ46260.1 CH477473 EAT40369.1 JTDY01001376 KOB74033.1 DS232416 EDS41481.1 CVRI01000015 CRK90021.1 KQ976509 KYM82672.1 ADTU01000292 GGFL01014087 MBW78265.1 GGFJ01008035 MBW57176.1 GANO01003865 JAB56006.1 GECU01003938 JAT03769.1 KZ288320 PBC28261.1 NNAY01001167 OXU24909.1 GFDL01008688 JAV26357.1 GEHC01000191 JAV47454.1 GEBQ01006703 JAT33274.1 JXUM01014438 KQ560403 KXJ82587.1 UFQT01000292 SSX22899.1 GEDC01023818 JAS13480.1 GGFM01005307 MBW26058.1 GEZM01007671 JAV95006.1 GECL01000248 JAP05876.1 GBGD01002260 JAC86629.1 JRES01000835 KNC27915.1 AJWK01019121 AJWK01019122 AJWK01019123 AJWK01019124 AJWK01019125 GBBI01002968 JAC15744.1 CM000160 EDW98848.1 KQ971371 EFA09998.1 CH940650 EDW67767.2 CH902617 EDV43831.1 AM231261 CAJ77194.1 AM231259 CAJ77192.1 CH933806 EDW16464.2 AM231258 CAJ77191.1 GAMC01003458 JAC03098.1 AM231256 CAJ77189.1 GAKP01005933 JAC53019.1 QCYY01002184 ROT72262.1 GDHF01030700 JAI21614.1 CH964272 EDW84562.1 CH916658 EDV90849.1 GBXI01011672 JAD02620.1 OUUW01000005 SPP80350.1 GDRN01100639 JAI58531.1 DS235797 EEB17207.1 CP012526 ALC46447.1 GDRN01100640 GDRN01100637 JAI58533.1 ACPB03010972 CM000070 EAL27617.1 GBHO01005244 GBHO01005243 GBHO01005242 GBHO01005241 GDHC01020271 JAG38360.1 JAG38361.1 JAG38362.1 JAG38363.1 JAP98357.1 JH431742 GBRD01008652 GBRD01008651 JAG57169.1 JXJN01019088 CH479179 EDW24916.1 AM231260 CAJ77193.1 AE014297 BT044519 AAF56377.1 ACH95293.1 CAEY01001944 AB091099 BAC22695.1 APGK01050286 KB741169 KB632294 ENN73307.1 ERL91664.1 AM231257 CH954182 CAJ77190.1 EDV53764.1 FN568102 CBH40223.1
SOQ49970.1 GDQN01010334 JAT80720.1 AGBW02011361 OWR46785.1 BABH01016664 KQ460952 KPJ10365.1 PCG63132.1 KZ150188 PZC72434.1 GL449414 EFN82787.1 NEVH01025129 PNF15988.1 KK852895 KDR14189.1 GGFK01007651 MBW40972.1 KQ760478 OAD60206.1 KQ977151 KYN05280.1 KQ981606 KYN39589.1 KQ980895 KYN11764.1 AXCM01002140 KK107250 QOIP01000002 EZA54390.1 RLU25841.1 GL888609 EGI59057.1 KQ983238 KYQ46260.1 CH477473 EAT40369.1 JTDY01001376 KOB74033.1 DS232416 EDS41481.1 CVRI01000015 CRK90021.1 KQ976509 KYM82672.1 ADTU01000292 GGFL01014087 MBW78265.1 GGFJ01008035 MBW57176.1 GANO01003865 JAB56006.1 GECU01003938 JAT03769.1 KZ288320 PBC28261.1 NNAY01001167 OXU24909.1 GFDL01008688 JAV26357.1 GEHC01000191 JAV47454.1 GEBQ01006703 JAT33274.1 JXUM01014438 KQ560403 KXJ82587.1 UFQT01000292 SSX22899.1 GEDC01023818 JAS13480.1 GGFM01005307 MBW26058.1 GEZM01007671 JAV95006.1 GECL01000248 JAP05876.1 GBGD01002260 JAC86629.1 JRES01000835 KNC27915.1 AJWK01019121 AJWK01019122 AJWK01019123 AJWK01019124 AJWK01019125 GBBI01002968 JAC15744.1 CM000160 EDW98848.1 KQ971371 EFA09998.1 CH940650 EDW67767.2 CH902617 EDV43831.1 AM231261 CAJ77194.1 AM231259 CAJ77192.1 CH933806 EDW16464.2 AM231258 CAJ77191.1 GAMC01003458 JAC03098.1 AM231256 CAJ77189.1 GAKP01005933 JAC53019.1 QCYY01002184 ROT72262.1 GDHF01030700 JAI21614.1 CH964272 EDW84562.1 CH916658 EDV90849.1 GBXI01011672 JAD02620.1 OUUW01000005 SPP80350.1 GDRN01100639 JAI58531.1 DS235797 EEB17207.1 CP012526 ALC46447.1 GDRN01100640 GDRN01100637 JAI58533.1 ACPB03010972 CM000070 EAL27617.1 GBHO01005244 GBHO01005243 GBHO01005242 GBHO01005241 GDHC01020271 JAG38360.1 JAG38361.1 JAG38362.1 JAG38363.1 JAP98357.1 JH431742 GBRD01008652 GBRD01008651 JAG57169.1 JXJN01019088 CH479179 EDW24916.1 AM231260 CAJ77193.1 AE014297 BT044519 AAF56377.1 ACH95293.1 CAEY01001944 AB091099 BAC22695.1 APGK01050286 KB741169 KB632294 ENN73307.1 ERL91664.1 AM231257 CH954182 CAJ77190.1 EDV53764.1 FN568102 CBH40223.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000008237
+ More
UP000235965 UP000027135 UP000075882 UP000078542 UP000075902 UP000075880 UP000078541 UP000078492 UP000075883 UP000053097 UP000279307 UP000007755 UP000075809 UP000008820 UP000037510 UP000002320 UP000183832 UP000078540 UP000075884 UP000005205 UP000242457 UP000215335 UP000005203 UP000002358 UP000069940 UP000249989 UP000037069 UP000092461 UP000075881 UP000075885 UP000002282 UP000007266 UP000095301 UP000008792 UP000007801 UP000009192 UP000192221 UP000283509 UP000007798 UP000001070 UP000268350 UP000009046 UP000095300 UP000092553 UP000015103 UP000069272 UP000001819 UP000075901 UP000092460 UP000008744 UP000092443 UP000078200 UP000000803 UP000015104 UP000092445 UP000019118 UP000030742 UP000008711
UP000235965 UP000027135 UP000075882 UP000078542 UP000075902 UP000075880 UP000078541 UP000078492 UP000075883 UP000053097 UP000279307 UP000007755 UP000075809 UP000008820 UP000037510 UP000002320 UP000183832 UP000078540 UP000075884 UP000005205 UP000242457 UP000215335 UP000005203 UP000002358 UP000069940 UP000249989 UP000037069 UP000092461 UP000075881 UP000075885 UP000002282 UP000007266 UP000095301 UP000008792 UP000007801 UP000009192 UP000192221 UP000283509 UP000007798 UP000001070 UP000268350 UP000009046 UP000095300 UP000092553 UP000015103 UP000069272 UP000001819 UP000075901 UP000092460 UP000008744 UP000092443 UP000078200 UP000000803 UP000015104 UP000092445 UP000019118 UP000030742 UP000008711
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JMP2
A0A2A4IVI1
A0A3S2LU13
A0A2H1WA90
A0A1E1W143
A0A212EZ58
+ More
H9JMP3 A0A0N1ICK8 A0A2A4IVC0 A0A2W1BFI1 E2BNK8 A0A2J7PI40 A0A067QVV9 A0A182L5R8 A0A2M4AJQ8 A0A310SK71 A0A151IKZ1 A0A182TTQ5 A0A182J539 A0A195FHX2 A0A151IVK2 A0A182M291 A0A026WE58 F4X376 A0A151WEJ6 Q170H3 A0A0L7LEQ1 B0X6J8 A0A1J1HPQ6 A0A195BEQ9 A0A182NE87 A0A158NLH9 A0A2M4DL65 A0A1S4FHV9 A0A2M4BVR8 U5ER90 A0A1B6JX31 A0A2A3EB24 A0A232F317 A0A088AQ60 K7J8T5 A0A1Q3FFM7 A0A1W7R8I2 A0A1B6MBL0 A0A182GN55 A0A336M1M5 A0A1B6CJ92 A0A2M3ZC79 A0A1Y1NFZ7 A0A0V0GD08 A0A069DYJ9 A0A0L0C6M0 A0A1B0CMR6 A0A023F434 A0A182K169 A0A182PB48 B4PUZ9 D6X0P7 A0A1I8NF56 B4LX20 B3LWZ2 Q2HPN8 Q2HPP0 B4K7U3 A0A1W4VL56 Q2HPP1 W8BV94 Q2HPP3 A0A034WEA3 A0A3R7MXU4 A0A0K8U4J1 B4NH65 B4K0Z5 A0A0A1WVS4 A0A3B0JXX2 A0A0P4VSM5 E0VV01 A0A1I8NP27 A0A0M4EQ88 A0A0P4VXD1 T1HLB7 A0A182F4E9 Q299T4 A0A0A9Z975 A0A182T624 T1J0Y7 A0A0K8SVC7 A0A1B0BRG9 B4G5Q0 A0A1A9YK92 A0A1A9UN89 Q2HPN9 Q9VBZ9 T1KB34 Q8I6J0 A0A1B0AAB3 N6U3Z6 Q2HPP2 D1H0P4
H9JMP3 A0A0N1ICK8 A0A2A4IVC0 A0A2W1BFI1 E2BNK8 A0A2J7PI40 A0A067QVV9 A0A182L5R8 A0A2M4AJQ8 A0A310SK71 A0A151IKZ1 A0A182TTQ5 A0A182J539 A0A195FHX2 A0A151IVK2 A0A182M291 A0A026WE58 F4X376 A0A151WEJ6 Q170H3 A0A0L7LEQ1 B0X6J8 A0A1J1HPQ6 A0A195BEQ9 A0A182NE87 A0A158NLH9 A0A2M4DL65 A0A1S4FHV9 A0A2M4BVR8 U5ER90 A0A1B6JX31 A0A2A3EB24 A0A232F317 A0A088AQ60 K7J8T5 A0A1Q3FFM7 A0A1W7R8I2 A0A1B6MBL0 A0A182GN55 A0A336M1M5 A0A1B6CJ92 A0A2M3ZC79 A0A1Y1NFZ7 A0A0V0GD08 A0A069DYJ9 A0A0L0C6M0 A0A1B0CMR6 A0A023F434 A0A182K169 A0A182PB48 B4PUZ9 D6X0P7 A0A1I8NF56 B4LX20 B3LWZ2 Q2HPN8 Q2HPP0 B4K7U3 A0A1W4VL56 Q2HPP1 W8BV94 Q2HPP3 A0A034WEA3 A0A3R7MXU4 A0A0K8U4J1 B4NH65 B4K0Z5 A0A0A1WVS4 A0A3B0JXX2 A0A0P4VSM5 E0VV01 A0A1I8NP27 A0A0M4EQ88 A0A0P4VXD1 T1HLB7 A0A182F4E9 Q299T4 A0A0A9Z975 A0A182T624 T1J0Y7 A0A0K8SVC7 A0A1B0BRG9 B4G5Q0 A0A1A9YK92 A0A1A9UN89 Q2HPN9 Q9VBZ9 T1KB34 Q8I6J0 A0A1B0AAB3 N6U3Z6 Q2HPP2 D1H0P4
PDB
4LW6
E-value=1.07349e-84,
Score=797
Ontologies
PATHWAY
GO
PANTHER
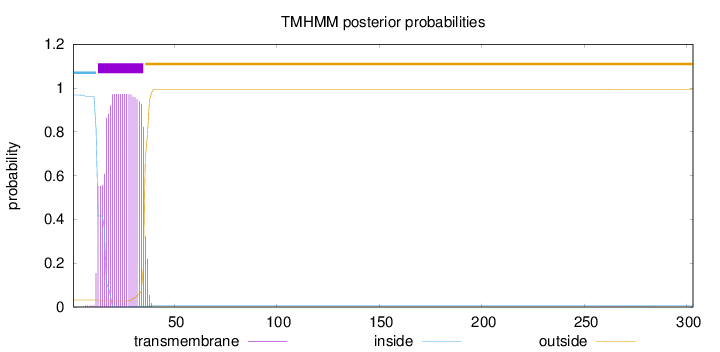
Topology
Length:
303
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.0312099999999
Exp number, first 60 AAs:
21.00671
Total prob of N-in:
0.96778
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 303
Population Genetic Test Statistics
Pi
324.507022
Theta
203.794709
Tajima's D
1.656712
CLR
0.028026
CSRT
0.822658867056647
Interpretation
Uncertain
