Gene
KWMTBOMO15521
Pre Gene Modal
BGIBMGA010749
Annotation
empty_spiracles_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 2.565
Sequence
CDS
ATGATGCCTCTCGCACCGACCCCTGTGGTGGCGGTGCCATCGAAGCCGAAAATCGGATTCAGCATAGACAGTATCGTTGGCGGTTCTGATAGGTCGCGGCAGGCGTCGCCGCCGCTGGCGAGCCCCGGGTCTCCGTCGCCGGTTGCCTCTCCGAGGAGTCCCTCGCCGCGTCCCCTGCTCCGGCCGAGCGCTTTGCCCGCCGTGTTCCCATCTCCGGAGCTGAAGAGGCTTCCGTACCTCGACGCCCAGAGTCACCACCATCAGTTCTTGGCGCATTTCCAAGGAGCGGCGCTGGCGGCAGCTTTAGCCCATCAGCAGCAAGGTTTCGGTGCCCCCCTACCGCCTCACCCGCCGCCGCACCCCGCCGGGCCGGTACCCAGAGAGTCTTACCAGCTCTACCCCTGGCTGATCAACAGGCACGGAAGGTTGTTCCCGCACAGGTTTCCTGGAGGTAAGCAGTTTCCAAACACTAATTAA
Protein
MMPLAPTPVVAVPSKPKIGFSIDSIVGGSDRSRQASPPLASPGSPSPVASPRSPSPRPLLRPSALPAVFPSPELKRLPYLDAQSHHHQFLAHFQGAALAAALAHQQQGFGAPLPPHPPPHPAGPVPRESYQLYPWLINRHGRLFPHRFPGGKQFPNTN
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
Ontologies
Topology
Subcellular location
Nucleus
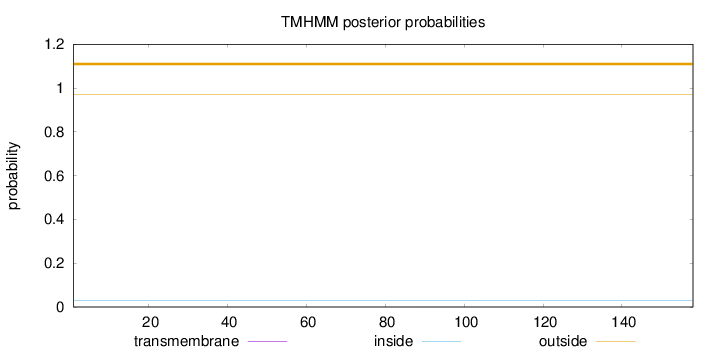
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00334
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.02857
outside
1 - 158
Population Genetic Test Statistics
Pi
230.839452
Theta
187.338749
Tajima's D
0.711037
CLR
0
CSRT
0.583220838958052
Interpretation
Uncertain
