Gene
KWMTBOMO15519
Pre Gene Modal
BGIBMGA010752
Annotation
PREDICTED:_tak1_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.264
Sequence
CDS
ATGATAGCAAGACCGTGGACTGACGATCTTCCGAAGTGCAGGAATACGTGTGTAGCTTCGTATTTTTCAAAGCTCGGAGGACACTCAAATGCTAACGATGATTATTTATGTTTTTTCTGTCAAACAGAGGATAACTTTTGTTTATATGGAGTATTTGAGCCACACAATGGTCTTGAAGCGGCCAGGCTTATCATGCAAAGAATGGCAGCTGAAATATTATTTCCACCACCCTCAGCCAACACAGATGAAGAGATACGAGACAGACTTAGGAATGCCTTTGTGTCAGTTGAGAAAGCATACATTGAAAACTATGATGGAATGATAGCAGAGAGAACTAGTTTGCAATATCAGCTGCAGACTTCTAGTGAGACTCAGAATCGCCAAATTATATTGTCTCGCTTAAAAGAAATAGACCAGCATCTGTCCGGAGGTGCTACTGTGGTCGTAGCTTTAGTGCACAAAAATAAACTATTTGTTGCTAATGTTGGTGAAAGTAGAGCTCTGCTATGTCGTACGGATGATAACTCGGTATTACGGGTCGTGCAGCTGACCGTTGACCACAGTCTCAACAACGAAGACGAGCTGCTCAGACTGTCTCAGCTAGGCTTAGACGTTAATAAACTGAGGAATGCGCAATACCTCGGCAACCAAACCGGGACCAGATGTCTGGGGAATTATTTAGTCAAGGGCCTGTACAAAGCGTTTCCCAGCATCAGCCCGGCCTCGTCCGAGCCCGTGGTGGCCTCCCCGGAGATACACGGACCCATCGTCCTGGACGACAGCTGTAGATTTCTGATCCTAGTAAGCGCGGGCGTTTACAAACGCATACAGGAGTCGAGGGGCAGCAGCGAACAAACGAACAAACAGCTCGCCCAGATGATCGTTGAGCACTTCAGAAAGCAGACAGACTTCAAGGCCGTCAGCGAAACGGTCCTGCACGAGATCGAGGAGGAGTTCATATCCCACTGCGTCAAGAACAACTTGACGCCCAAATCGAACACTCACGTCACGCTGCTCATAAGGAACTTTAACTTTATTGCGGCCGCGGGGGCGGAGCCCGGGCCTCCCAGGAGCGCCTCGGTGCGCTTCAACCCGGTCGTGCAGTCCAAGTCCAGCACGCTCTTAGATGAGATGGAGTCCAGCTCGCCGGAGTCCCCGGTCGACACGTGCCCGGGCCGCCAGTACGACAAGCACGTGCGCATCAAGGGCTACGTTGACTTCGCGTGTTACTACGAGAACGTGGAAAGGGCGCGTCGCGGCGGCACCCTGCCAGACTTCATCCAGTGA
Protein
MIARPWTDDLPKCRNTCVASYFSKLGGHSNANDDYLCFFCQTEDNFCLYGVFEPHNGLEAARLIMQRMAAEILFPPPSANTDEEIRDRLRNAFVSVEKAYIENYDGMIAERTSLQYQLQTSSETQNRQIILSRLKEIDQHLSGGATVVVALVHKNKLFVANVGESRALLCRTDDNSVLRVVQLTVDHSLNNEDELLRLSQLGLDVNKLRNAQYLGNQTGTRCLGNYLVKGLYKAFPSISPASSEPVVASPEIHGPIVLDDSCRFLILVSAGVYKRIQESRGSSEQTNKQLAQMIVEHFRKQTDFKAVSETVLHEIEEEFISHCVKNNLTPKSNTHVTLLIRNFNFIAAAGAEPGPPRSASVRFNPVVQSKSSTLLDEMESSSPESPVDTCPGRQYDKHVRIKGYVDFACYYENVERARRGGTLPDFIQ
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the PP2C family.
Uniprot
E9JEI9
A0A2A4JFE8
A0A0N1PIH9
A0A194PK24
A0A212FHC2
A0A2W1B521
+ More
E2AP93 F4WLE1 A0A195BMY3 A0A151WMI9 A0A158NVJ6 A0A154P4A2 A0A2J7RFM0 E2BNJ6 A0A195CTB6 A0A151IQX6 A0A0C9RJS8 A0A195FY30 A0A026X3X4 A0A232FG71 A0A067QIQ1 A0A3L8DKP7 E9J703 A0A0J7NTH7 A0A2A3EDN1 A0A088ADH0 A0A1W4XJV0 K7IU64 A0A1B6DES7 A0A0L7RJM4 A0A1S3CUL0 A0A1Y1LV76 E0VTI6 A0A2J7RFL9 J3JT92 N6SYL1 A0A0M8ZX79 A0A2P8YLB4 D6X284 A0A1B6L8I0 A0A1B6FLQ9 A0A0P5TK51 A0A023F253 A0A0A9X5Y3 A0A0P6JQL7 A0A1D2NLT8 A0A0K8S543 A0A0P6E3F0 A0A0A9XE84 A0A2L2YHH8 A0A0P5U4N4 A0A2J7RFL6 A0A0P5ITR2 T1HNA7 A0A0P5J2K7 A0A1L8DKN1 A0A0P5TU61 A0A0P5XEM6 A0A1B0GHI4 A0A0K8S3X3 A0A0P4ZNY0 A0A0P6BQ63 A0A0N8D2D0 A0A0K8TR94 A0A0P5IKK0 A0A2S2QA86 A0A1B6IWQ7 E9G673 A0A0P4VKX9 A0A0N8BWS2 A0A0P5UV92 A0A0P6F9D5 A0A0P5E0L0 A0A210QNM7 A0A2C9K3A5 A0A2C9K3B7 A0A1B0EYZ2 A0A226DF56 V4CH24 A0A2R2V0G6 A0A3R7N5V9 A0A0P4W6Y8 A0A0P4W3T6 A0A2S2NW81 A0A182UHP5 B0W3C4 A0A182VK83 Q7QD46 A0A224XJE5 A0A182JPW7 A0A0B6ZFE9 A0A2Y9QBH0 J9JZ99 A0A0B6ZF35 D4A6C6
E2AP93 F4WLE1 A0A195BMY3 A0A151WMI9 A0A158NVJ6 A0A154P4A2 A0A2J7RFM0 E2BNJ6 A0A195CTB6 A0A151IQX6 A0A0C9RJS8 A0A195FY30 A0A026X3X4 A0A232FG71 A0A067QIQ1 A0A3L8DKP7 E9J703 A0A0J7NTH7 A0A2A3EDN1 A0A088ADH0 A0A1W4XJV0 K7IU64 A0A1B6DES7 A0A0L7RJM4 A0A1S3CUL0 A0A1Y1LV76 E0VTI6 A0A2J7RFL9 J3JT92 N6SYL1 A0A0M8ZX79 A0A2P8YLB4 D6X284 A0A1B6L8I0 A0A1B6FLQ9 A0A0P5TK51 A0A023F253 A0A0A9X5Y3 A0A0P6JQL7 A0A1D2NLT8 A0A0K8S543 A0A0P6E3F0 A0A0A9XE84 A0A2L2YHH8 A0A0P5U4N4 A0A2J7RFL6 A0A0P5ITR2 T1HNA7 A0A0P5J2K7 A0A1L8DKN1 A0A0P5TU61 A0A0P5XEM6 A0A1B0GHI4 A0A0K8S3X3 A0A0P4ZNY0 A0A0P6BQ63 A0A0N8D2D0 A0A0K8TR94 A0A0P5IKK0 A0A2S2QA86 A0A1B6IWQ7 E9G673 A0A0P4VKX9 A0A0N8BWS2 A0A0P5UV92 A0A0P6F9D5 A0A0P5E0L0 A0A210QNM7 A0A2C9K3A5 A0A2C9K3B7 A0A1B0EYZ2 A0A226DF56 V4CH24 A0A2R2V0G6 A0A3R7N5V9 A0A0P4W6Y8 A0A0P4W3T6 A0A2S2NW81 A0A182UHP5 B0W3C4 A0A182VK83 Q7QD46 A0A224XJE5 A0A182JPW7 A0A0B6ZFE9 A0A2Y9QBH0 J9JZ99 A0A0B6ZF35 D4A6C6
Pubmed
19121390
21040523
26354079
22118469
28756777
20798317
+ More
21719571 21347285 24508170 28648823 24845553 30249741 21282665 20075255 28004739 20566863 22516182 23537049 29403074 18362917 19820115 25474469 25401762 26823975 27289101 26561354 26369729 21292972 27129103 28812685 15562597 23254933 12364791 14747013 17210077 18058037 15057822
21719571 21347285 24508170 28648823 24845553 30249741 21282665 20075255 28004739 20566863 22516182 23537049 29403074 18362917 19820115 25474469 25401762 26823975 27289101 26561354 26369729 21292972 27129103 28812685 15562597 23254933 12364791 14747013 17210077 18058037 15057822
EMBL
BABH01016642
GQ426306
ADM32532.1
NWSH01001651
PCG70539.1
KQ460952
+ More
KPJ10369.1 KQ459601 KPI93672.1 AGBW02008505 OWR53141.1 KZ150330 PZC71359.1 GL441481 EFN64766.1 GL888207 EGI65104.1 KQ976433 KYM87348.1 KQ982944 KYQ49040.1 ADTU01027228 KQ434803 KZC05950.1 NEVH01004410 PNF39626.1 GL449414 EFN82775.1 KQ977349 KYN03374.1 KQ981155 KYN08849.1 GBYB01008455 JAG78222.1 KQ981169 KYN45348.1 KK107019 EZA62793.1 NNAY01000282 OXU29520.1 KK853305 KDR08718.1 QOIP01000007 RLU20951.1 GL768421 EFZ11379.1 LBMM01001801 KMQ95725.1 KZ288271 PBC29837.1 AAZX01002661 GEDC01013094 JAS24204.1 KQ414581 KOC70998.1 GEZM01046122 JAV77499.1 DS235767 EEB16713.1 PNF39628.1 BT126448 AEE61412.1 APGK01058789 APGK01058790 KB741292 ENN70303.1 KQ435812 KOX72757.1 PYGN01000512 PSN45036.1 KQ971371 EFA09896.1 GEBQ01020011 JAT19966.1 GECZ01018631 JAS51138.1 GDIP01130744 LRGB01000512 JAL72970.1 KZS18750.1 GBBI01003556 JAC15156.1 GBHO01028195 GDHC01008447 JAG15409.1 JAQ10182.1 GDIQ01000877 JAN93860.1 LJIJ01000009 ODN06222.1 GBRD01017972 JAG47855.1 GDIQ01069563 JAN25174.1 GBHO01028194 GDHC01014172 GDHC01009656 JAG15410.1 JAQ04457.1 JAQ08973.1 IAAA01021520 LAA07578.1 GDIP01119688 JAL84026.1 PNF39627.1 GDIQ01233799 JAK17926.1 ACPB03007651 GDIQ01230398 JAK21327.1 GFDF01007066 JAV07018.1 GDIP01139547 JAL64167.1 GDIP01074210 JAM29505.1 AJWK01005182 GBRD01017973 JAG47854.1 GDIP01223237 JAJ00165.1 GDIP01015090 JAM88625.1 GDIP01075607 JAM28108.1 GDAI01000704 JAI16899.1 GDIQ01214412 JAK37313.1 GGMS01005338 MBY74541.1 GECU01016339 JAS91367.1 GL732533 EFX85043.1 GDKW01003420 JAI53175.1 GDIQ01137283 JAL14443.1 GDIP01109045 JAL94669.1 GDIQ01066471 JAN28266.1 GDIP01148295 JAJ75107.1 NEDP02002665 OWF50308.1 AJVK01033072 LNIX01000020 OXA44182.1 KB200521 ESP01385.1 KY683840 QCYY01002775 AQY45916.1 ROT67702.1 QCYY01001430 ROT78099.1 GDRN01087047 JAI61086.1 GDRN01087050 GDRN01087049 GDRN01087045 JAI61084.1 GGMR01008367 MBY20986.1 DS231831 EDS31267.1 AAAB01008859 EAA07598.5 GFTR01005252 JAW11174.1 HACG01020287 CEK67152.1 ABLF02028255 HACG01020288 CEK67153.1 AC127784
KPJ10369.1 KQ459601 KPI93672.1 AGBW02008505 OWR53141.1 KZ150330 PZC71359.1 GL441481 EFN64766.1 GL888207 EGI65104.1 KQ976433 KYM87348.1 KQ982944 KYQ49040.1 ADTU01027228 KQ434803 KZC05950.1 NEVH01004410 PNF39626.1 GL449414 EFN82775.1 KQ977349 KYN03374.1 KQ981155 KYN08849.1 GBYB01008455 JAG78222.1 KQ981169 KYN45348.1 KK107019 EZA62793.1 NNAY01000282 OXU29520.1 KK853305 KDR08718.1 QOIP01000007 RLU20951.1 GL768421 EFZ11379.1 LBMM01001801 KMQ95725.1 KZ288271 PBC29837.1 AAZX01002661 GEDC01013094 JAS24204.1 KQ414581 KOC70998.1 GEZM01046122 JAV77499.1 DS235767 EEB16713.1 PNF39628.1 BT126448 AEE61412.1 APGK01058789 APGK01058790 KB741292 ENN70303.1 KQ435812 KOX72757.1 PYGN01000512 PSN45036.1 KQ971371 EFA09896.1 GEBQ01020011 JAT19966.1 GECZ01018631 JAS51138.1 GDIP01130744 LRGB01000512 JAL72970.1 KZS18750.1 GBBI01003556 JAC15156.1 GBHO01028195 GDHC01008447 JAG15409.1 JAQ10182.1 GDIQ01000877 JAN93860.1 LJIJ01000009 ODN06222.1 GBRD01017972 JAG47855.1 GDIQ01069563 JAN25174.1 GBHO01028194 GDHC01014172 GDHC01009656 JAG15410.1 JAQ04457.1 JAQ08973.1 IAAA01021520 LAA07578.1 GDIP01119688 JAL84026.1 PNF39627.1 GDIQ01233799 JAK17926.1 ACPB03007651 GDIQ01230398 JAK21327.1 GFDF01007066 JAV07018.1 GDIP01139547 JAL64167.1 GDIP01074210 JAM29505.1 AJWK01005182 GBRD01017973 JAG47854.1 GDIP01223237 JAJ00165.1 GDIP01015090 JAM88625.1 GDIP01075607 JAM28108.1 GDAI01000704 JAI16899.1 GDIQ01214412 JAK37313.1 GGMS01005338 MBY74541.1 GECU01016339 JAS91367.1 GL732533 EFX85043.1 GDKW01003420 JAI53175.1 GDIQ01137283 JAL14443.1 GDIP01109045 JAL94669.1 GDIQ01066471 JAN28266.1 GDIP01148295 JAJ75107.1 NEDP02002665 OWF50308.1 AJVK01033072 LNIX01000020 OXA44182.1 KB200521 ESP01385.1 KY683840 QCYY01002775 AQY45916.1 ROT67702.1 QCYY01001430 ROT78099.1 GDRN01087047 JAI61086.1 GDRN01087050 GDRN01087049 GDRN01087045 JAI61084.1 GGMR01008367 MBY20986.1 DS231831 EDS31267.1 AAAB01008859 EAA07598.5 GFTR01005252 JAW11174.1 HACG01020287 CEK67152.1 ABLF02028255 HACG01020288 CEK67153.1 AC127784
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000000311
+ More
UP000007755 UP000078540 UP000075809 UP000005205 UP000076502 UP000235965 UP000008237 UP000078542 UP000078492 UP000078541 UP000053097 UP000215335 UP000027135 UP000279307 UP000036403 UP000242457 UP000005203 UP000192223 UP000002358 UP000053825 UP000079169 UP000009046 UP000019118 UP000053105 UP000245037 UP000007266 UP000076858 UP000094527 UP000015103 UP000092461 UP000000305 UP000242188 UP000076420 UP000092462 UP000198287 UP000030746 UP000283509 UP000075902 UP000002320 UP000075903 UP000007062 UP000075881 UP000248483 UP000007819 UP000002494
UP000007755 UP000078540 UP000075809 UP000005205 UP000076502 UP000235965 UP000008237 UP000078542 UP000078492 UP000078541 UP000053097 UP000215335 UP000027135 UP000279307 UP000036403 UP000242457 UP000005203 UP000192223 UP000002358 UP000053825 UP000079169 UP000009046 UP000019118 UP000053105 UP000245037 UP000007266 UP000076858 UP000094527 UP000015103 UP000092461 UP000000305 UP000242188 UP000076420 UP000092462 UP000198287 UP000030746 UP000283509 UP000075902 UP000002320 UP000075903 UP000007062 UP000075881 UP000248483 UP000007819 UP000002494
Interpro
SUPFAM
SSF81606
SSF81606
Gene 3D
CDD
ProteinModelPortal
E9JEI9
A0A2A4JFE8
A0A0N1PIH9
A0A194PK24
A0A212FHC2
A0A2W1B521
+ More
E2AP93 F4WLE1 A0A195BMY3 A0A151WMI9 A0A158NVJ6 A0A154P4A2 A0A2J7RFM0 E2BNJ6 A0A195CTB6 A0A151IQX6 A0A0C9RJS8 A0A195FY30 A0A026X3X4 A0A232FG71 A0A067QIQ1 A0A3L8DKP7 E9J703 A0A0J7NTH7 A0A2A3EDN1 A0A088ADH0 A0A1W4XJV0 K7IU64 A0A1B6DES7 A0A0L7RJM4 A0A1S3CUL0 A0A1Y1LV76 E0VTI6 A0A2J7RFL9 J3JT92 N6SYL1 A0A0M8ZX79 A0A2P8YLB4 D6X284 A0A1B6L8I0 A0A1B6FLQ9 A0A0P5TK51 A0A023F253 A0A0A9X5Y3 A0A0P6JQL7 A0A1D2NLT8 A0A0K8S543 A0A0P6E3F0 A0A0A9XE84 A0A2L2YHH8 A0A0P5U4N4 A0A2J7RFL6 A0A0P5ITR2 T1HNA7 A0A0P5J2K7 A0A1L8DKN1 A0A0P5TU61 A0A0P5XEM6 A0A1B0GHI4 A0A0K8S3X3 A0A0P4ZNY0 A0A0P6BQ63 A0A0N8D2D0 A0A0K8TR94 A0A0P5IKK0 A0A2S2QA86 A0A1B6IWQ7 E9G673 A0A0P4VKX9 A0A0N8BWS2 A0A0P5UV92 A0A0P6F9D5 A0A0P5E0L0 A0A210QNM7 A0A2C9K3A5 A0A2C9K3B7 A0A1B0EYZ2 A0A226DF56 V4CH24 A0A2R2V0G6 A0A3R7N5V9 A0A0P4W6Y8 A0A0P4W3T6 A0A2S2NW81 A0A182UHP5 B0W3C4 A0A182VK83 Q7QD46 A0A224XJE5 A0A182JPW7 A0A0B6ZFE9 A0A2Y9QBH0 J9JZ99 A0A0B6ZF35 D4A6C6
E2AP93 F4WLE1 A0A195BMY3 A0A151WMI9 A0A158NVJ6 A0A154P4A2 A0A2J7RFM0 E2BNJ6 A0A195CTB6 A0A151IQX6 A0A0C9RJS8 A0A195FY30 A0A026X3X4 A0A232FG71 A0A067QIQ1 A0A3L8DKP7 E9J703 A0A0J7NTH7 A0A2A3EDN1 A0A088ADH0 A0A1W4XJV0 K7IU64 A0A1B6DES7 A0A0L7RJM4 A0A1S3CUL0 A0A1Y1LV76 E0VTI6 A0A2J7RFL9 J3JT92 N6SYL1 A0A0M8ZX79 A0A2P8YLB4 D6X284 A0A1B6L8I0 A0A1B6FLQ9 A0A0P5TK51 A0A023F253 A0A0A9X5Y3 A0A0P6JQL7 A0A1D2NLT8 A0A0K8S543 A0A0P6E3F0 A0A0A9XE84 A0A2L2YHH8 A0A0P5U4N4 A0A2J7RFL6 A0A0P5ITR2 T1HNA7 A0A0P5J2K7 A0A1L8DKN1 A0A0P5TU61 A0A0P5XEM6 A0A1B0GHI4 A0A0K8S3X3 A0A0P4ZNY0 A0A0P6BQ63 A0A0N8D2D0 A0A0K8TR94 A0A0P5IKK0 A0A2S2QA86 A0A1B6IWQ7 E9G673 A0A0P4VKX9 A0A0N8BWS2 A0A0P5UV92 A0A0P6F9D5 A0A0P5E0L0 A0A210QNM7 A0A2C9K3A5 A0A2C9K3B7 A0A1B0EYZ2 A0A226DF56 V4CH24 A0A2R2V0G6 A0A3R7N5V9 A0A0P4W6Y8 A0A0P4W3T6 A0A2S2NW81 A0A182UHP5 B0W3C4 A0A182VK83 Q7QD46 A0A224XJE5 A0A182JPW7 A0A0B6ZFE9 A0A2Y9QBH0 J9JZ99 A0A0B6ZF35 D4A6C6
PDB
2J4O
E-value=3.59261e-53,
Score=527
Ontologies
GO
PANTHER
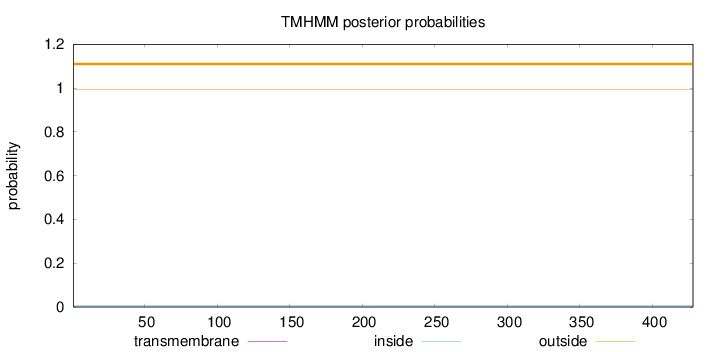
Topology
Length:
428
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00189
Exp number, first 60 AAs:
0.00031
Total prob of N-in:
0.00541
outside
1 - 428
Population Genetic Test Statistics
Pi
240.162466
Theta
213.96434
Tajima's D
0.434426
CLR
19.312654
CSRT
0.496325183740813
Interpretation
Uncertain
