Gene
KWMTBOMO15518 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010794
Annotation
26S_proteasome_regulatory_ATPase_subunit_10B_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.999
Sequence
CDS
ATGCCGGCTAGTACATCGGACATGGAGCCGTTAAGAGAAAAGGCCTTCCAGGATTACAGAAAGAAGCTCATGGAGCATAAGGAAGTCGAGTCACGACTCAAAGAAGGTCGTGACCAACTAAAAGATTTGACCAAACAATATGACAAGAGTGAAAATGACCTGAAAGCCTTACAAAGTGTTGGACAAATAGTTGGAGAAGTCTTGAAACAGCTCACAGAAGAAAAATTTATTGTAAAAGCTACTAATGGCCCTCGCTACGTTGTTGGCTGTCGTCGGCAACTTGACAAGAACAAGCTTAAAGGAGGAACACGAGTTGCCCTCGACATGACTACTCTTACAATAATGAGACATCTGCCTAGAGAGGTTGATCCCCTAGTTTACAACATGAGTCATGAGGATCCTGGTGACGTGACCTATTCCGCCATAGGAGGTCTTCAAGAACAGATTAGACAGCTGAGAGAGGTGATTGAACTTCCCCTAATGAACCCAGAGCTGTTTGTTCGAGTCGGTATCACTCCACCCAAAGGATGTTTGCTGTACGGACCTCCTGGTACAGGAAAGACTTTGCTCGCGAGGGCTGTGGCTTCACAGCTCGATGCTAACTTTTTAAAGGTGGTGTCATCGGCGATTGTTGACAAGTACATCGGTGAATCGGCCCGCCTCATCCGGGAGATGTTCAACTACGCGCGGGACCACCAACCCTGCATCATATTCATGGACGAGATCGATGCCATCGGTGGGAGACGTTTTTCCGAAGGCACGAGCGCCGATCGAGAGATCCAACGTACCCTGATGGAGTTGTTGAACCAAATGGATGGTTTCGACTCCTTGGGACAGGTGAAGATCATCATGGCCACCAACAGACCAGACACGCTGGACCCCGCTCTGCTGAGGCCCGGAAGATTGGACAGGAAAATCGAAATACCCCTGCCCAATGAACAGGCTAGATTGGAAATCTTGAAGATCCACGCGTCGCCGATCGCCAAGCACGGCGAGATGGACTACGAGGCGGTCGTGAAGCTTTCAGATACTTTCAACGGCGCGGACCTGCGCAACGTCTGCACGGAGGCAGGACTCTTCGCCATCAGGGCCGAGCGGGAGTACATCATACAGGAGGATCTAATGAAGGCCGTCCGTAAAGTAGCCGACAACAAAAAGCTGGAGAGCAAACTGGACTACAAGCCCGTCTGA
Protein
MPASTSDMEPLREKAFQDYRKKLMEHKEVESRLKEGRDQLKDLTKQYDKSENDLKALQSVGQIVGEVLKQLTEEKFIVKATNGPRYVVGCRRQLDKNKLKGGTRVALDMTTLTIMRHLPREVDPLVYNMSHEDPGDVTYSAIGGLQEQIRQLREVIELPLMNPELFVRVGITPPKGCLLYGPPGTGKTLLARAVASQLDANFLKVVSSAIVDKYIGESARLIREMFNYARDHQPCIIFMDEIDAIGGRRFSEGTSADREIQRTLMELLNQMDGFDSLGQVKIIMATNRPDTLDPALLRPGRLDRKIEIPLPNEQARLEILKIHASPIAKHGEMDYEAVVKLSDTFNGADLRNVCTEAGLFAIRAEREYIIQEDLMKAVRKVADNKKLESKLDYKPV
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
Q1HPL0
I4DLA7
A0A2A4JFR0
A0A2W1B6Z9
A0A3S2M8K9
O62556
+ More
A0A212FHC6 S4PFJ3 A0A2H1WKA2 D7EKM4 E1ZW28 A0A1B6JZB9 A0A1B6DIC1 A0A0C9R062 A0A0J7KZQ6 A0A1W4WU39 A0A3L8DBU0 A0A0B4J2R5 A0A336K1E2 A0A2J7R9A3 A0A1Y1MJI7 E2BHF7 T1IGG6 A0A1B0D1K5 A0A1B6MIQ2 A0A069DU17 A0A151JR92 A0A195D407 F4W5K5 A0A151I373 A0A1Q3G308 A0A158NSC6 A0A151X6Z1 B0W8M6 Q1HQM1 A0A023ETJ7 A0A1L8DSX4 A0A0P6B2J0 A0A1B0CQJ7 A0A067R5C0 A0A182RNG2 A0A2A3EV07 R4FP59 A0A0B4J2P2 J3JV20 A0A2M4AE32 A0A0A9W076 A0A154PLX3 A0A195ERZ5 A0A182P2G1 A0A2M3Z931 A0A2M4BQX0 W5JLL0 A0A182F7A3 A0A182M1H8 A0A182YGG7 A0A182VR97 A0A182SND6 U5EWD3 A0A0P4VQM4 A0A0V0G5P9 J3JW58 A0A182VBX6 A0A182JPC7 A0A182TS40 A0A182L932 A0A182X9U4 Q7PXH7 A0A182HKS6 A0A182QMG1 A0A164K4A0 A0A0L0BTK4 A0A182N5E1 A0A182J259 A0A0M4EJ03 B4JWS6 E9H6I3 A0A084W575 B4MA11 A0A023FB11 A0A1A9X5B2 A0A232ELP1 B4L6T8 A0A1L8ECZ0 T1PCR9 A0A1I8PUH6 B4NER3 W8BQZ7 A0A0K8WEU5 A0A0C9R8S6 A0A034V6R2 A0A1W4VF45 H9JMP1 A0A1B0FKF3 A0A1A9XE03 A0A1A9VRW4 A0A1B0AN55 A0A194PJR5 B3MRC7 A0A1A9ZIF7
A0A212FHC6 S4PFJ3 A0A2H1WKA2 D7EKM4 E1ZW28 A0A1B6JZB9 A0A1B6DIC1 A0A0C9R062 A0A0J7KZQ6 A0A1W4WU39 A0A3L8DBU0 A0A0B4J2R5 A0A336K1E2 A0A2J7R9A3 A0A1Y1MJI7 E2BHF7 T1IGG6 A0A1B0D1K5 A0A1B6MIQ2 A0A069DU17 A0A151JR92 A0A195D407 F4W5K5 A0A151I373 A0A1Q3G308 A0A158NSC6 A0A151X6Z1 B0W8M6 Q1HQM1 A0A023ETJ7 A0A1L8DSX4 A0A0P6B2J0 A0A1B0CQJ7 A0A067R5C0 A0A182RNG2 A0A2A3EV07 R4FP59 A0A0B4J2P2 J3JV20 A0A2M4AE32 A0A0A9W076 A0A154PLX3 A0A195ERZ5 A0A182P2G1 A0A2M3Z931 A0A2M4BQX0 W5JLL0 A0A182F7A3 A0A182M1H8 A0A182YGG7 A0A182VR97 A0A182SND6 U5EWD3 A0A0P4VQM4 A0A0V0G5P9 J3JW58 A0A182VBX6 A0A182JPC7 A0A182TS40 A0A182L932 A0A182X9U4 Q7PXH7 A0A182HKS6 A0A182QMG1 A0A164K4A0 A0A0L0BTK4 A0A182N5E1 A0A182J259 A0A0M4EJ03 B4JWS6 E9H6I3 A0A084W575 B4MA11 A0A023FB11 A0A1A9X5B2 A0A232ELP1 B4L6T8 A0A1L8ECZ0 T1PCR9 A0A1I8PUH6 B4NER3 W8BQZ7 A0A0K8WEU5 A0A0C9R8S6 A0A034V6R2 A0A1W4VF45 H9JMP1 A0A1B0FKF3 A0A1A9XE03 A0A1A9VRW4 A0A1B0AN55 A0A194PJR5 B3MRC7 A0A1A9ZIF7
Pubmed
22651552
28756777
10464306
22118469
23622113
18362917
+ More
19820115 20798317 30249741 20075255 28004739 26334808 21719571 21347285 17204158 17510324 24945155 26483478 24845553 22516182 25401762 26823975 20920257 23761445 25244985 27129103 20966253 12364791 14747013 17210077 26108605 17994087 21292972 24438588 25474469 28648823 25315136 24495485 25348373 19121390 26354079
19820115 20798317 30249741 20075255 28004739 26334808 21719571 21347285 17204158 17510324 24945155 26483478 24845553 22516182 25401762 26823975 20920257 23761445 25244985 27129103 20966253 12364791 14747013 17210077 26108605 17994087 21292972 24438588 25474469 28648823 25315136 24495485 25348373 19121390 26354079
EMBL
DQ443392
ABF51481.1
AK402075
BAM18697.1
NWSH01001651
PCG70538.1
+ More
KZ150330 PZC71358.1 RSAL01000008 RVE53946.1 AJ223384 CAA11285.1 AGBW02008505 OWR53142.1 GAIX01003992 JAA88568.1 ODYU01009127 SOQ53332.1 DS497757 EFA13218.1 GL434758 EFN74615.1 GECU01003166 JAT04541.1 GEDC01011855 JAS25443.1 GBYB01006312 JAG76079.1 LBMM01001759 KMQ95803.1 QOIP01000010 RLU17368.1 UFQS01000040 UFQT01000040 SSW98277.1 SSX18663.1 NEVH01006600 PNF37411.1 GEZM01029915 GEZM01029913 JAV85881.1 GL448287 EFN84926.1 ACPB03001111 AJVK01010278 GEBQ01004186 JAT35791.1 GBGD01001702 JAC87187.1 KQ978642 KYN29497.1 KQ976881 KYN07581.1 GL887655 EGI70502.1 KQ976543 KYM80982.1 GFDL01000862 JAV34183.1 ADTU01024816 KQ982459 KYQ56141.1 DS231859 EDS39076.1 DQ440423 CH477308 ABF18456.1 EAT44043.1 JXUM01143234 GAPW01001874 KQ569558 JAC11724.1 KXJ68590.1 GFDF01004573 JAV09511.1 GDIP01022269 JAM81446.1 AJWK01023689 KK852954 KDR13296.1 KZ288185 PBC34979.1 GAHY01001049 JAA76461.1 BT127086 AEE62048.1 GGFK01005723 MBW39044.1 GBHO01045334 GBRD01000237 GDHC01002380 JAF98269.1 JAG65584.1 JAQ16249.1 KQ434977 KZC12833.1 KQ981993 KYN31025.1 GGFM01004282 MBW25033.1 GGFJ01006232 MBW55373.1 ADMH02001070 ETN64193.1 AXCM01003332 GANO01001472 JAB58399.1 GDKW01001280 JAI55315.1 GECL01003437 JAP02687.1 BT127476 AEE62438.1 AAAB01008987 EAA01092.5 APCN01000860 AXCN02001178 LRGB01003375 KZS02938.1 JRES01001480 KNC22559.1 CP012528 ALC49004.1 CH916376 EDV95202.1 GL732597 EFX72665.1 ATLV01020513 KE525302 KFB45369.1 CH940655 EDW66070.1 GBBI01000594 JAC18118.1 NNAY01003522 OXU19267.1 CH933812 EDW06084.1 GFDG01002198 JAV16601.1 KA646546 AFP61175.1 CH964239 EDW82232.1 GAMC01005093 JAC01463.1 GDHF01002692 JAI49622.1 GBYB01003191 JAG72958.1 GAKP01021736 GAKP01021734 GAKP01021732 GAKP01021731 JAC37218.1 BABH01016642 CCAG010007411 JXJN01000725 KQ459601 KPI93671.1 CH902622 EDV34332.1
KZ150330 PZC71358.1 RSAL01000008 RVE53946.1 AJ223384 CAA11285.1 AGBW02008505 OWR53142.1 GAIX01003992 JAA88568.1 ODYU01009127 SOQ53332.1 DS497757 EFA13218.1 GL434758 EFN74615.1 GECU01003166 JAT04541.1 GEDC01011855 JAS25443.1 GBYB01006312 JAG76079.1 LBMM01001759 KMQ95803.1 QOIP01000010 RLU17368.1 UFQS01000040 UFQT01000040 SSW98277.1 SSX18663.1 NEVH01006600 PNF37411.1 GEZM01029915 GEZM01029913 JAV85881.1 GL448287 EFN84926.1 ACPB03001111 AJVK01010278 GEBQ01004186 JAT35791.1 GBGD01001702 JAC87187.1 KQ978642 KYN29497.1 KQ976881 KYN07581.1 GL887655 EGI70502.1 KQ976543 KYM80982.1 GFDL01000862 JAV34183.1 ADTU01024816 KQ982459 KYQ56141.1 DS231859 EDS39076.1 DQ440423 CH477308 ABF18456.1 EAT44043.1 JXUM01143234 GAPW01001874 KQ569558 JAC11724.1 KXJ68590.1 GFDF01004573 JAV09511.1 GDIP01022269 JAM81446.1 AJWK01023689 KK852954 KDR13296.1 KZ288185 PBC34979.1 GAHY01001049 JAA76461.1 BT127086 AEE62048.1 GGFK01005723 MBW39044.1 GBHO01045334 GBRD01000237 GDHC01002380 JAF98269.1 JAG65584.1 JAQ16249.1 KQ434977 KZC12833.1 KQ981993 KYN31025.1 GGFM01004282 MBW25033.1 GGFJ01006232 MBW55373.1 ADMH02001070 ETN64193.1 AXCM01003332 GANO01001472 JAB58399.1 GDKW01001280 JAI55315.1 GECL01003437 JAP02687.1 BT127476 AEE62438.1 AAAB01008987 EAA01092.5 APCN01000860 AXCN02001178 LRGB01003375 KZS02938.1 JRES01001480 KNC22559.1 CP012528 ALC49004.1 CH916376 EDV95202.1 GL732597 EFX72665.1 ATLV01020513 KE525302 KFB45369.1 CH940655 EDW66070.1 GBBI01000594 JAC18118.1 NNAY01003522 OXU19267.1 CH933812 EDW06084.1 GFDG01002198 JAV16601.1 KA646546 AFP61175.1 CH964239 EDW82232.1 GAMC01005093 JAC01463.1 GDHF01002692 JAI49622.1 GBYB01003191 JAG72958.1 GAKP01021736 GAKP01021734 GAKP01021732 GAKP01021731 JAC37218.1 BABH01016642 CCAG010007411 JXJN01000725 KQ459601 KPI93671.1 CH902622 EDV34332.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000007266
UP000000311
UP000036403
+ More
UP000192223 UP000279307 UP000002358 UP000235965 UP000008237 UP000015103 UP000092462 UP000078492 UP000078542 UP000007755 UP000078540 UP000005205 UP000075809 UP000002320 UP000008820 UP000069940 UP000249989 UP000092461 UP000027135 UP000075900 UP000242457 UP000005203 UP000076502 UP000078541 UP000075885 UP000000673 UP000069272 UP000075883 UP000076408 UP000075920 UP000075901 UP000075903 UP000075881 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075886 UP000076858 UP000037069 UP000075884 UP000075880 UP000092553 UP000001070 UP000000305 UP000030765 UP000008792 UP000091820 UP000215335 UP000009192 UP000095301 UP000095300 UP000007798 UP000192221 UP000005204 UP000092444 UP000092443 UP000078200 UP000092460 UP000053268 UP000007801 UP000092445
UP000192223 UP000279307 UP000002358 UP000235965 UP000008237 UP000015103 UP000092462 UP000078492 UP000078542 UP000007755 UP000078540 UP000005205 UP000075809 UP000002320 UP000008820 UP000069940 UP000249989 UP000092461 UP000027135 UP000075900 UP000242457 UP000005203 UP000076502 UP000078541 UP000075885 UP000000673 UP000069272 UP000075883 UP000076408 UP000075920 UP000075901 UP000075903 UP000075881 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075886 UP000076858 UP000037069 UP000075884 UP000075880 UP000092553 UP000001070 UP000000305 UP000030765 UP000008792 UP000091820 UP000215335 UP000009192 UP000095301 UP000095300 UP000007798 UP000192221 UP000005204 UP000092444 UP000092443 UP000078200 UP000092460 UP000053268 UP000007801 UP000092445
Interpro
ProteinModelPortal
Q1HPL0
I4DLA7
A0A2A4JFR0
A0A2W1B6Z9
A0A3S2M8K9
O62556
+ More
A0A212FHC6 S4PFJ3 A0A2H1WKA2 D7EKM4 E1ZW28 A0A1B6JZB9 A0A1B6DIC1 A0A0C9R062 A0A0J7KZQ6 A0A1W4WU39 A0A3L8DBU0 A0A0B4J2R5 A0A336K1E2 A0A2J7R9A3 A0A1Y1MJI7 E2BHF7 T1IGG6 A0A1B0D1K5 A0A1B6MIQ2 A0A069DU17 A0A151JR92 A0A195D407 F4W5K5 A0A151I373 A0A1Q3G308 A0A158NSC6 A0A151X6Z1 B0W8M6 Q1HQM1 A0A023ETJ7 A0A1L8DSX4 A0A0P6B2J0 A0A1B0CQJ7 A0A067R5C0 A0A182RNG2 A0A2A3EV07 R4FP59 A0A0B4J2P2 J3JV20 A0A2M4AE32 A0A0A9W076 A0A154PLX3 A0A195ERZ5 A0A182P2G1 A0A2M3Z931 A0A2M4BQX0 W5JLL0 A0A182F7A3 A0A182M1H8 A0A182YGG7 A0A182VR97 A0A182SND6 U5EWD3 A0A0P4VQM4 A0A0V0G5P9 J3JW58 A0A182VBX6 A0A182JPC7 A0A182TS40 A0A182L932 A0A182X9U4 Q7PXH7 A0A182HKS6 A0A182QMG1 A0A164K4A0 A0A0L0BTK4 A0A182N5E1 A0A182J259 A0A0M4EJ03 B4JWS6 E9H6I3 A0A084W575 B4MA11 A0A023FB11 A0A1A9X5B2 A0A232ELP1 B4L6T8 A0A1L8ECZ0 T1PCR9 A0A1I8PUH6 B4NER3 W8BQZ7 A0A0K8WEU5 A0A0C9R8S6 A0A034V6R2 A0A1W4VF45 H9JMP1 A0A1B0FKF3 A0A1A9XE03 A0A1A9VRW4 A0A1B0AN55 A0A194PJR5 B3MRC7 A0A1A9ZIF7
A0A212FHC6 S4PFJ3 A0A2H1WKA2 D7EKM4 E1ZW28 A0A1B6JZB9 A0A1B6DIC1 A0A0C9R062 A0A0J7KZQ6 A0A1W4WU39 A0A3L8DBU0 A0A0B4J2R5 A0A336K1E2 A0A2J7R9A3 A0A1Y1MJI7 E2BHF7 T1IGG6 A0A1B0D1K5 A0A1B6MIQ2 A0A069DU17 A0A151JR92 A0A195D407 F4W5K5 A0A151I373 A0A1Q3G308 A0A158NSC6 A0A151X6Z1 B0W8M6 Q1HQM1 A0A023ETJ7 A0A1L8DSX4 A0A0P6B2J0 A0A1B0CQJ7 A0A067R5C0 A0A182RNG2 A0A2A3EV07 R4FP59 A0A0B4J2P2 J3JV20 A0A2M4AE32 A0A0A9W076 A0A154PLX3 A0A195ERZ5 A0A182P2G1 A0A2M3Z931 A0A2M4BQX0 W5JLL0 A0A182F7A3 A0A182M1H8 A0A182YGG7 A0A182VR97 A0A182SND6 U5EWD3 A0A0P4VQM4 A0A0V0G5P9 J3JW58 A0A182VBX6 A0A182JPC7 A0A182TS40 A0A182L932 A0A182X9U4 Q7PXH7 A0A182HKS6 A0A182QMG1 A0A164K4A0 A0A0L0BTK4 A0A182N5E1 A0A182J259 A0A0M4EJ03 B4JWS6 E9H6I3 A0A084W575 B4MA11 A0A023FB11 A0A1A9X5B2 A0A232ELP1 B4L6T8 A0A1L8ECZ0 T1PCR9 A0A1I8PUH6 B4NER3 W8BQZ7 A0A0K8WEU5 A0A0C9R8S6 A0A034V6R2 A0A1W4VF45 H9JMP1 A0A1B0FKF3 A0A1A9XE03 A0A1A9VRW4 A0A1B0AN55 A0A194PJR5 B3MRC7 A0A1A9ZIF7
PDB
5T0C
E-value=0,
Score=1738
Ontologies
GO
PANTHER
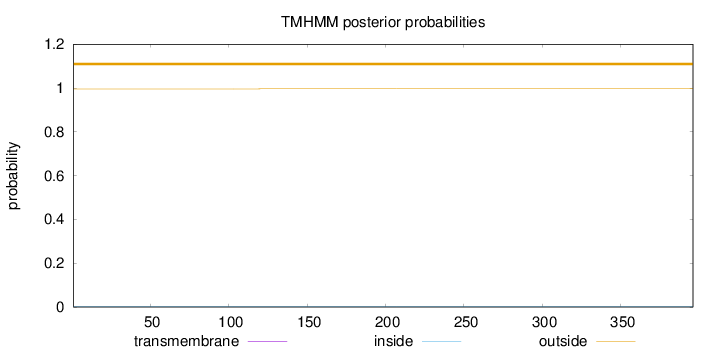
Topology
Length:
396
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00103
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00382
outside
1 - 396
Population Genetic Test Statistics
Pi
267.825179
Theta
156.319846
Tajima's D
1.97127
CLR
0.195157
CSRT
0.875656217189141
Interpretation
Uncertain
