Gene
KWMTBOMO15517
Annotation
hypothetical_protein_M514_27221_[Trichuris_suis]
Location in the cell
Mitochondrial Reliability : 1.421 Nuclear Reliability : 1.532
Sequence
CDS
ATGTGGTACGAATATTTTCCTAATATTACACCTATTGAATGGGCTACACCCACAGTTAACATTCTGAAATCTACAGGGGAAATTAGGATATGTGGAGACTACAGAACTACACTTAATCCAGTACTTATTAAACATCTACACCCAGTTCCGATTTTCGATCAACTACGCCAAAATCTCGCAAACGGTAAATTATTTTCAAAAATAGACCTTAAAGACGCATACTTACAATTTGAAATAGCACCAGATTCGAAAAAGTACTTGACGTTGTCAACCCATAAAGGGTATTTCCAATACAATAGAATGCCTTTTGGAATATCGACTGCTCCTTCAATTTTTCAACATTTTCTGGATCAACTGCTAGGCGATATCACCAATGTAGCTGTATATTTTGACGACATAGCTATAGCAGGGAAAGATCTTTCAGAACATTTACAAACTTTATCTATTGTATTCGATCGCTTACAAAACGCTGGCCTAAAAGTTAATTTAAAGAAATATACAAAGAAATGCATAGCTTCTTCACAACCATTAATTGCATTTGACGAGAAACGTCCCCTTTATTTAGCATGTGACGCTTCCGAAAAGGGATTGGGTGCAGTATTATTCCACAAGGACTCGAACATAGAACAGCCAATTGCTTTTGCTTCAAGGAAACTACGACCAGCGGAAATGAAATATTCCGTTATTGATCGTGAAGCTTTAGCAATAGTATTTGGTATAAAAAAATTTGATCAGTACTTGAGAGGAACGAAATTTAATCTAGTTACGGACCATAAGCCTCTTATACATATTATGGGTGCACGCCGCAACCTTCCTAAACTAGCCAATAACCGATTGGTAAGATGGGCCCTAGTAGTGGGAAGTTATCAATATGACATTTACTACAGGAAAGGTGAAAATAACACATTAGCTGATTGCCTTTCCAGATTACCAAACCCTGAAACTGAACCCTCCGAGACAGAAGGGCTTGTACATAAAATAGATCTACGTCTTTTGAGCACTCGAATGACTGACCTTAACCTTTCAGAACAGTTACTTATGAAAACAACTGCAAAGGATCATATACTCACAAAAGTTTGTCAAAATTTAAAAACCGGCTGGAGAGAATCAGAATACAATCCAGAGATGAAACCATTCTACAGAAACCGGACTGAATTATCTGTTGAGAATAAGATACTTATGAGGCAAGGACGCATCGTTATACCTACAGCACTCCGAAAAGCCATTCTTACATACCTTCATCGAGGACACCCTGGCATTTCTGCTATGAAAGCACTTTCACGTTACTATGTTTGGTGGCCTAACCTTGACGAAGACATAGAATTATTTGTCAAGAAATGTACCAGATGCCAACAGAACCGCCCTTGTAATCCTGAACTTCCTGTATTTTCCTGGTCCATACCAGAAGAAGTATGGGAGAGAATCCATATCGACTTTGCTGGACCGTTCGAAGGATCGTATTGGTTGGTATTGTGCGACGCTCTCTCCAAATGGGTGGAAATACGACCGATGAAACACATCAACACCAGATCACTTTGTCTTACATTAGATAACATATTTTGTACATTTGGCTTACCAAAAATGATTATATCCGATAATGGACCCCAATTTACATCTTATGAATTTAAAGAATATTGCACAAAACAGTCTATTTTACATGTCACATCATCCCCATATCATCCTCGGACAAATGGGCTGGCAGAACGTTTAGTCAGAACATTCAAAAATAGAATGGCATCGGTAGACAACACAAATCTCGAGCGCCGACTATTAGAATTTCTGTTTACATACAGAAACACTCCGCACTCATCCACTGGTAAATCTCCAGCTGAGATGATGTTTGGAAGACAATTGAATTGCATACTTTCCAACATTCGGCCAGACAAAAGAAGGTTAATGCAGTATTTACAAGTAAAAGAAAATATAATAATGATAGGTAGCAAAAATGACAATGTGTTTGAAGTATATTTCAAATAA
Protein
MWYEYFPNITPIEWATPTVNILKSTGEIRICGDYRTTLNPVLIKHLHPVPIFDQLRQNLANGKLFSKIDLKDAYLQFEIAPDSKKYLTLSTHKGYFQYNRMPFGISTAPSIFQHFLDQLLGDITNVAVYFDDIAIAGKDLSEHLQTLSIVFDRLQNAGLKVNLKKYTKKCIASSQPLIAFDEKRPLYLACDASEKGLGAVLFHKDSNIEQPIAFASRKLRPAEMKYSVIDREALAIVFGIKKFDQYLRGTKFNLVTDHKPLIHIMGARRNLPKLANNRLVRWALVVGSYQYDIYYRKGENNTLADCLSRLPNPETEPSETEGLVHKIDLRLLSTRMTDLNLSEQLLMKTTAKDHILTKVCQNLKTGWRESEYNPEMKPFYRNRTELSVENKILMRQGRIVIPTALRKAILTYLHRGHPGISAMKALSRYYVWWPNLDEDIELFVKKCTRCQQNRPCNPELPVFSWSIPEEVWERIHIDFAGPFEGSYWLVLCDALSKWVEIRPMKHINTRSLCLTLDNIFCTFGLPKMIISDNGPQFTSYEFKEYCTKQSILHVTSSPYHPRTNGLAERLVRTFKNRMASVDNTNLERRLLEFLFTYRNTPHSSTGKSPAEMMFGRQLNCILSNIRPDKRRLMQYLQVKENIIMIGSKNDNVFEVYFK
Summary
Uniprot
A0A085MTP2
A0A0A9W826
A0A0A9W3I3
A0A2B4S0C2
A0A085MRU5
A0A146L9N5
+ More
A0A224XI32 A0A085M406 A0A0C2JXZ5 A0A3P8REG6 A0A183BQV4 A0A0A9XIJ3 A0A0V0RWN6 A0A3P8NQC4 A0A0C2JFK2 A0A3B3B4L9 A0A0C2IHZ5 A0A3P8NEQ7 A0A3P8NV61 A0A0C2IU42 A0A1X7T719 A0A1X7STP0 A0A3B3HJL6 W4LMH6 A0A3B5BG14 A0A3P8Q506 A0A3B3TFN7 A0A147BII2 A0A1X7TWV5 A0A3Q1F281 A0A131XQT5 I3KY30 A0A1S3L075 A0A1E1XN56 A0A1X7VU64 A0A0V0WST9 A0A0V1M0R9 A0A0V0ZE23 A0A0V0RTT5 A0A3Q1GLI5 A0A147BIP4 W4Z9D5 A0A3B3H2N7 A0A2G8LRL0 A0A1X7TIE8 A2TGR5 A0A0V1C3T1 A0A085MQC7 A0A0V0VH72 A0A0V0US17 I3KYR6 A0A1A7WUZ7 A0A0V1KNI7 A0A131Y536 A0A147BJB1 A0A0V0USW3 A0A1X7U623 A0A0V1BVI1 A0A1A7YSF0 A0A2B4RSM3 A2TGR4 A0A0V1FKD1 A0A3P8VSE7 T2MJH3 A0A267G4Y1 A0A267H2K6 A0A2G8LB01 A0A1A8QZH2 A0A085N023 A0A090XCC9 A0A3R7DF32 A0A1S3SSU6 A0A0A9XFS1 A0A3Q1EQC9 A0A2B4S2I1 A0A1S3KHX3 A0A1B6LWG3 L7LTN5 A0A0V1CV78 A0A147BJE5 A0A146LWN6 A0A085LJR8 A0A0V0VH28 A0A0A9W0S7 A0A146NT21 A0A0V1DDI4 A0A147BK15 A0A267H2J3 A0A146XMZ2 A0A2B4R6J0 A0A1S3J391 A0A2R2MK13 A0A147BMH4 A0A0V1A810 A0A3P8QD94 A0A267F772
A0A224XI32 A0A085M406 A0A0C2JXZ5 A0A3P8REG6 A0A183BQV4 A0A0A9XIJ3 A0A0V0RWN6 A0A3P8NQC4 A0A0C2JFK2 A0A3B3B4L9 A0A0C2IHZ5 A0A3P8NEQ7 A0A3P8NV61 A0A0C2IU42 A0A1X7T719 A0A1X7STP0 A0A3B3HJL6 W4LMH6 A0A3B5BG14 A0A3P8Q506 A0A3B3TFN7 A0A147BII2 A0A1X7TWV5 A0A3Q1F281 A0A131XQT5 I3KY30 A0A1S3L075 A0A1E1XN56 A0A1X7VU64 A0A0V0WST9 A0A0V1M0R9 A0A0V0ZE23 A0A0V0RTT5 A0A3Q1GLI5 A0A147BIP4 W4Z9D5 A0A3B3H2N7 A0A2G8LRL0 A0A1X7TIE8 A2TGR5 A0A0V1C3T1 A0A085MQC7 A0A0V0VH72 A0A0V0US17 I3KYR6 A0A1A7WUZ7 A0A0V1KNI7 A0A131Y536 A0A147BJB1 A0A0V0USW3 A0A1X7U623 A0A0V1BVI1 A0A1A7YSF0 A0A2B4RSM3 A2TGR4 A0A0V1FKD1 A0A3P8VSE7 T2MJH3 A0A267G4Y1 A0A267H2K6 A0A2G8LB01 A0A1A8QZH2 A0A085N023 A0A090XCC9 A0A3R7DF32 A0A1S3SSU6 A0A0A9XFS1 A0A3Q1EQC9 A0A2B4S2I1 A0A1S3KHX3 A0A1B6LWG3 L7LTN5 A0A0V1CV78 A0A147BJE5 A0A146LWN6 A0A085LJR8 A0A0V0VH28 A0A0A9W0S7 A0A146NT21 A0A0V1DDI4 A0A147BK15 A0A267H2J3 A0A146XMZ2 A0A2B4R6J0 A0A1S3J391 A0A2R2MK13 A0A147BMH4 A0A0V1A810 A0A3P8QD94 A0A267F772
Pubmed
EMBL
KL367659
KFD60588.1
GBHO01042589
JAG01015.1
GBHO01042596
JAG01008.1
+ More
LSMT01000238 PFX22483.1 KL367712 KFD59941.1 GDHC01014693 GDHC01004534 JAQ03936.1 JAQ14095.1 GFTR01008725 JAW07701.1 KL363234 KFD51952.1 JWZT01000439 KII74358.1 GBHO01045651 GBHO01045644 GBHO01026684 GBHO01026679 JAF97952.1 JAF97959.1 JAG16920.1 JAG16925.1 JYDL01000067 KRX18868.1 JWZT01002984 KII68048.1 JWZT01004052 KII64969.1 JWZT01005674 KII60352.1 AZHX01001899 ETW98900.1 GEGO01004818 JAR90586.1 GEFM01006397 JAP69399.1 AERX01052799 AERX01052800 GFAA01002662 JAU00773.1 JYDK01000060 KRX78865.1 JYDO01000377 KRZ65389.1 JYDQ01000223 KRY10622.1 JYDL01000080 KRX17894.1 GEGO01004800 JAR90604.1 AAGJ04101977 MRZV01000005 PIK62810.1 EF199622 ABM90393.1 JYDI01000800 KRY43965.1 KL367976 KFD59423.1 JYDN01000036 KRX62859.1 JYDN01000181 KRX54086.1 AERX01039690 HADW01008367 SBP09767.1 JYDW01000348 KRZ48953.1 GEFM01002211 JAP73585.1 GEGO01004546 JAR90858.1 KRX54093.1 JYDH01000010 KRY40903.1 HADX01010638 SBP32870.1 LSMT01000343 PFX19819.1 EF199621 ABM90392.1 JYDT01000070 KRY86520.1 HAAD01006009 CDG72241.1 NIVC01000551 PAA81070.1 NIVC01000050 PAA92506.1 MRZV01000143 PIK57404.1 HAEH01013823 SBR98409.1 KL367587 KFD62819.1 GBIH01000761 JAC93949.1 NIRI01001153 RJW67559.1 GBHO01027659 JAG15945.1 LSMT01000232 PFX22695.1 GEBQ01020673 GEBQ01011946 JAT19304.1 JAT28031.1 GACK01009588 JAA55446.1 JYDI01000102 KRY52595.1 GEGO01004812 JAR90592.1 GDHC01007869 JAQ10760.1 KL363640 KFD45214.1 KRX62850.1 GBHO01042583 JAG01021.1 GCES01151725 JAQ34597.1 JYDI01000010 KRY59721.1 GEGO01004729 JAR90675.1 NIVC01000075 PAA91739.1 GCES01042893 JAR43430.1 LSMT01001659 PFX12008.1 GEGO01003414 JAR91990.1 JYDQ01000025 KRY20467.1 NIVC01001308 PAA69606.1
LSMT01000238 PFX22483.1 KL367712 KFD59941.1 GDHC01014693 GDHC01004534 JAQ03936.1 JAQ14095.1 GFTR01008725 JAW07701.1 KL363234 KFD51952.1 JWZT01000439 KII74358.1 GBHO01045651 GBHO01045644 GBHO01026684 GBHO01026679 JAF97952.1 JAF97959.1 JAG16920.1 JAG16925.1 JYDL01000067 KRX18868.1 JWZT01002984 KII68048.1 JWZT01004052 KII64969.1 JWZT01005674 KII60352.1 AZHX01001899 ETW98900.1 GEGO01004818 JAR90586.1 GEFM01006397 JAP69399.1 AERX01052799 AERX01052800 GFAA01002662 JAU00773.1 JYDK01000060 KRX78865.1 JYDO01000377 KRZ65389.1 JYDQ01000223 KRY10622.1 JYDL01000080 KRX17894.1 GEGO01004800 JAR90604.1 AAGJ04101977 MRZV01000005 PIK62810.1 EF199622 ABM90393.1 JYDI01000800 KRY43965.1 KL367976 KFD59423.1 JYDN01000036 KRX62859.1 JYDN01000181 KRX54086.1 AERX01039690 HADW01008367 SBP09767.1 JYDW01000348 KRZ48953.1 GEFM01002211 JAP73585.1 GEGO01004546 JAR90858.1 KRX54093.1 JYDH01000010 KRY40903.1 HADX01010638 SBP32870.1 LSMT01000343 PFX19819.1 EF199621 ABM90392.1 JYDT01000070 KRY86520.1 HAAD01006009 CDG72241.1 NIVC01000551 PAA81070.1 NIVC01000050 PAA92506.1 MRZV01000143 PIK57404.1 HAEH01013823 SBR98409.1 KL367587 KFD62819.1 GBIH01000761 JAC93949.1 NIRI01001153 RJW67559.1 GBHO01027659 JAG15945.1 LSMT01000232 PFX22695.1 GEBQ01020673 GEBQ01011946 JAT19304.1 JAT28031.1 GACK01009588 JAA55446.1 JYDI01000102 KRY52595.1 GEGO01004812 JAR90592.1 GDHC01007869 JAQ10760.1 KL363640 KFD45214.1 KRX62850.1 GBHO01042583 JAG01021.1 GCES01151725 JAQ34597.1 JYDI01000010 KRY59721.1 GEGO01004729 JAR90675.1 NIVC01000075 PAA91739.1 GCES01042893 JAR43430.1 LSMT01001659 PFX12008.1 GEGO01003414 JAR91990.1 JYDQ01000025 KRY20467.1 NIVC01001308 PAA69606.1
Proteomes
UP000225706
UP000265100
UP000050741
UP000054630
UP000261560
UP000007879
+ More
UP000001038 UP000019140 UP000261400 UP000261540 UP000257200 UP000005207 UP000087266 UP000054673 UP000054843 UP000054783 UP000007110 UP000230750 UP000054653 UP000054681 UP000054721 UP000054776 UP000054995 UP000265120 UP000215902 UP000265000 UP000085678
UP000001038 UP000019140 UP000261400 UP000261540 UP000257200 UP000005207 UP000087266 UP000054673 UP000054843 UP000054783 UP000007110 UP000230750 UP000054653 UP000054681 UP000054721 UP000054776 UP000054995 UP000265120 UP000215902 UP000265000 UP000085678
Pfam
Interpro
IPR041588
Integrase_H2C2
+ More
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR021109 Peptidase_aspartic_dom_sf
IPR041373 RT_RNaseH
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR012341 6hp_glycosidase-like_sf
IPR020464 LanC-like_prot_euk
IPR007822 LANC-like
IPR034128 K02A2.6-like
IPR018061 Retropepsins
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR001969 Aspartic_peptidase_AS
IPR021109 Peptidase_aspartic_dom_sf
IPR041373 RT_RNaseH
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR012341 6hp_glycosidase-like_sf
IPR020464 LanC-like_prot_euk
IPR007822 LANC-like
IPR034128 K02A2.6-like
IPR018061 Retropepsins
Gene 3D
ProteinModelPortal
A0A085MTP2
A0A0A9W826
A0A0A9W3I3
A0A2B4S0C2
A0A085MRU5
A0A146L9N5
+ More
A0A224XI32 A0A085M406 A0A0C2JXZ5 A0A3P8REG6 A0A183BQV4 A0A0A9XIJ3 A0A0V0RWN6 A0A3P8NQC4 A0A0C2JFK2 A0A3B3B4L9 A0A0C2IHZ5 A0A3P8NEQ7 A0A3P8NV61 A0A0C2IU42 A0A1X7T719 A0A1X7STP0 A0A3B3HJL6 W4LMH6 A0A3B5BG14 A0A3P8Q506 A0A3B3TFN7 A0A147BII2 A0A1X7TWV5 A0A3Q1F281 A0A131XQT5 I3KY30 A0A1S3L075 A0A1E1XN56 A0A1X7VU64 A0A0V0WST9 A0A0V1M0R9 A0A0V0ZE23 A0A0V0RTT5 A0A3Q1GLI5 A0A147BIP4 W4Z9D5 A0A3B3H2N7 A0A2G8LRL0 A0A1X7TIE8 A2TGR5 A0A0V1C3T1 A0A085MQC7 A0A0V0VH72 A0A0V0US17 I3KYR6 A0A1A7WUZ7 A0A0V1KNI7 A0A131Y536 A0A147BJB1 A0A0V0USW3 A0A1X7U623 A0A0V1BVI1 A0A1A7YSF0 A0A2B4RSM3 A2TGR4 A0A0V1FKD1 A0A3P8VSE7 T2MJH3 A0A267G4Y1 A0A267H2K6 A0A2G8LB01 A0A1A8QZH2 A0A085N023 A0A090XCC9 A0A3R7DF32 A0A1S3SSU6 A0A0A9XFS1 A0A3Q1EQC9 A0A2B4S2I1 A0A1S3KHX3 A0A1B6LWG3 L7LTN5 A0A0V1CV78 A0A147BJE5 A0A146LWN6 A0A085LJR8 A0A0V0VH28 A0A0A9W0S7 A0A146NT21 A0A0V1DDI4 A0A147BK15 A0A267H2J3 A0A146XMZ2 A0A2B4R6J0 A0A1S3J391 A0A2R2MK13 A0A147BMH4 A0A0V1A810 A0A3P8QD94 A0A267F772
A0A224XI32 A0A085M406 A0A0C2JXZ5 A0A3P8REG6 A0A183BQV4 A0A0A9XIJ3 A0A0V0RWN6 A0A3P8NQC4 A0A0C2JFK2 A0A3B3B4L9 A0A0C2IHZ5 A0A3P8NEQ7 A0A3P8NV61 A0A0C2IU42 A0A1X7T719 A0A1X7STP0 A0A3B3HJL6 W4LMH6 A0A3B5BG14 A0A3P8Q506 A0A3B3TFN7 A0A147BII2 A0A1X7TWV5 A0A3Q1F281 A0A131XQT5 I3KY30 A0A1S3L075 A0A1E1XN56 A0A1X7VU64 A0A0V0WST9 A0A0V1M0R9 A0A0V0ZE23 A0A0V0RTT5 A0A3Q1GLI5 A0A147BIP4 W4Z9D5 A0A3B3H2N7 A0A2G8LRL0 A0A1X7TIE8 A2TGR5 A0A0V1C3T1 A0A085MQC7 A0A0V0VH72 A0A0V0US17 I3KYR6 A0A1A7WUZ7 A0A0V1KNI7 A0A131Y536 A0A147BJB1 A0A0V0USW3 A0A1X7U623 A0A0V1BVI1 A0A1A7YSF0 A0A2B4RSM3 A2TGR4 A0A0V1FKD1 A0A3P8VSE7 T2MJH3 A0A267G4Y1 A0A267H2K6 A0A2G8LB01 A0A1A8QZH2 A0A085N023 A0A090XCC9 A0A3R7DF32 A0A1S3SSU6 A0A0A9XFS1 A0A3Q1EQC9 A0A2B4S2I1 A0A1S3KHX3 A0A1B6LWG3 L7LTN5 A0A0V1CV78 A0A147BJE5 A0A146LWN6 A0A085LJR8 A0A0V0VH28 A0A0A9W0S7 A0A146NT21 A0A0V1DDI4 A0A147BK15 A0A267H2J3 A0A146XMZ2 A0A2B4R6J0 A0A1S3J391 A0A2R2MK13 A0A147BMH4 A0A0V1A810 A0A3P8QD94 A0A267F772
PDB
4OL8
E-value=1.0207e-20,
Score=249
Ontologies
GO
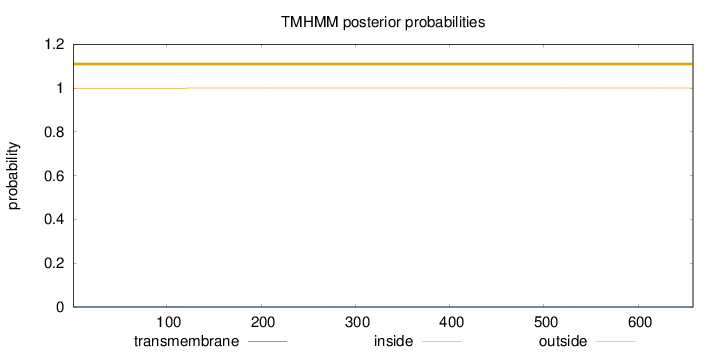
Topology
Length:
658
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00872999999999998
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.00068
outside
1 - 658
Population Genetic Test Statistics
Pi
239.819761
Theta
144.17596
Tajima's D
2.000867
CLR
11.637196
CSRT
0.87935603219839
Interpretation
Uncertain
