Pre Gene Modal
BGIBMGA010792
Annotation
PREDICTED:_nucleolar_protein_14_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 4.506
Sequence
CDS
ATGGCAAAAGTTAAAAACAAGCGGAATTCAGGACTAGCTGATAAAGTTCACAGTAATAGAAAAGCAGAGATCAACAAAAAGAAAATCAATCCATTTGAGGTGCATGTGAATCGTGAAAAAATTAAAGTTTTAGGGAAGAAATCGAAGCATGATAAAGGTTTGCCTGGTGTATCTCGAGCTAAAGCTGTACAGAAGAGAAAAGAGACATTGGGCACAGAAATGAAGCTGATGAATAAGACAAACACATTTATTGATCGTCGTATTGGTGAGAAAAACAACCAGTTGTCAGCTGAAGATAAAATGATTGCGAGATTTGCCGCTGAAAGAGTTAAACAACACAATAAAAAGAGTATATACAATTTGGCAGATGATGAGATATTAACACATAGAGGCCAAACGTTGGAGCAAATAGAAAAGTTTGATGATCCAAGATCTGATGATGAAGATGAAGAAGGAAAGGCATATGGTGGACTAGACTATGACTTTGTTGCTGAAGGCCACTTTGGTGGAGGAATGCTTTCAAAAACTGACTCAAAAGACGGTGCAAAATCTCACAAAGATTTAATAGAGCAGTTAATAGCAGAGTCAAAGAAAAGGAAGGCAGAGAAGCAGAAAGATAAAGAACAGACTTTAGATTTGACAGAAAAGTTAGATAGTGAATGGAAAGATCTTCAACCGGTTGTTTTTAAGAGACCGAGAGTAGAAGAAGAGTCTGCTATTGATAAAGTGCTGAATAAAGCAGAAAAGAATAGTCAGGATTATGATAAGATGATGAGAGAGTTGAAGTTTGAGAAGAGAGGAACGCCTTCAGATGCCCTAAAAAGCGACGAGAAACAAGCAAAAGAAGAGAGGGAAAAGTTAGAGCAGTGGGAGAGAGAACGACAACAGAGAATGACGGGAGATGATGAAGTAATACCTGTAAGGGTTAAACAACCTGCTCCTAAACACAGGTCAGCCGAGGACCTTGATGACAATTTCGAGTTGGAAGACGAGGCTAGTTTTATGTTGGCGTATGATAAGGATGGAAAACCTTTAGTACCAGAACATATTCTTAAAGATTATTCAGTACCAAACGCAAAACCGAAGTTTGAAGATGGCGTTTCGGATACGTCTGATGAAGAAAACAGTGATGATGCTGATGATGAAAGTAACGGTGATGATGATGGAGATAGCGATGAACAAGAAGTGTCGGATGAAGATGCACCTGAAGAGACTACTGATGATGAAAAACATGTCAACGATACCGACAACAAAGGTGCAGCAAAAGAAATAAATGATAACAGAGTTGAGGGAAAGAAACTTGAAGTTGAAAACCTTTCATCACAGTCTGAATCTGAAGAGGATACTGAAGATGAACTAAACGTAAATATAGAAGAGGGAACCAAAGTTAAAGAACCCCAAAAAGTTACAACCCAAATTCAAATTAATAATGTTGATACTTCAGAAGACAGCTCAAACGATATCCCAGAATCTAATGACAAACAGAATGTGCTTAAGATAACTGAAAAAAATTGTATATCAAATAATGTTTCAGCTAAATGTAATGGAAATGTAATTAATGAGAACAATCGTAAGAATGTGGATGATCTGAACGCTTTTAATGATTTGAAATCATCTAGTGATGAGAGTGATGACAGTGATGTAGATGATACAGAAATCGGCGCTATGTTTGAGAAGAGCAATGAAGGTGAAAGTTTAAAGGAATTATTGTCCGGAAAAACACCTTCACAGCAGTATACAGCCATACAGAAACTGATAAAAACATATGATCCCAGCTTAGGCGAGGAGAACAAAGACAAACTGAGTCGCCTATTCGCTCACTTACTACAACACATCAACGATATTTTTACCAATTTAAAAGACGAAGGCTCCGTTATAAAAGCCTTCCTGATATTCGATAGATTAGTACCTCATATGTTTGATCTGGCCAAGCTGAATAAAATTTCAACTAAAAAGTACTTGGTCGATCTGTTGGTAGAAAAGCGAAATAAATATATGAAACGCCCGAAACGTATACCGGATTTGGACACGTTAATATTTTTCAAATTGATCTCGCTGCTGTATCCCACGTCCGACTTTCGGCATCCGGTAACGACGCCATCTTTGTTGTTTATGTCCGAGATTTTGAATCGGTCTAGATTCGACGACGCTTTCTCGATATCAAGGGGATTCTTTGTGACAGCCCTAATTTTAGAATACACTGTTCTGTCAAAACGATTAGTACCGTCCGCGCTGAATTACTTAAGAGGCATTTTATATTATTGTGCGAACACATCAATACTTAATCAAGTACAATTAGTCCCGCCTTTTATGTTACATAAAATGGAGAAGATTCTAAATTTAAACACAGATTGTTCTAAAATGTCGGTCGCGAACAAGATGGCCGCTAAAGATTTGATTCTAGAGAGTGTAGATGATGACTTCAAAATCCGCTGTTTGCTGATCAGTACGGTGATGTTGAAGGAGTTCTTTGACAATTTTAATGAGTTAGAGGCACAGGAATGCATGTTTGAGTATCACATAAAGTTGTTAGCCAGGATCGATACAGAATTGTACCCCAAGAAGGTGGCCAGTGTCGTTTCCGAAGTAAGTCAGTACATGAAAGAGTGCTTAGAAATGAAAACTTACACTCCGTTGTGTAGAGAGAAAAAGATACCCAAAGCTTTGAGGTTGTATGAACCGGATGTGCAGGAGGTTTTCACTGGAAGTAAAGGCTCTAAATTATCAAGAGAACAGGCGGAAGCTGCTCGTTTGAAGACTAAACTAAAGAAAGAGATGAAGGGAGCTCTCCGTGAGATCAGAAGAGACAAGTCCTATATAGCTTCTGTTAAGCTAAAGGAGAAGATGCGCAGTGACAATATAAGAAAAGAAAAGGTAAAACAAATCTACAAAGAGGCGGCCATACAACAAGGAGAATTGAATAAGCTGAAGAGAAGTAAATGA
Protein
MAKVKNKRNSGLADKVHSNRKAEINKKKINPFEVHVNREKIKVLGKKSKHDKGLPGVSRAKAVQKRKETLGTEMKLMNKTNTFIDRRIGEKNNQLSAEDKMIARFAAERVKQHNKKSIYNLADDEILTHRGQTLEQIEKFDDPRSDDEDEEGKAYGGLDYDFVAEGHFGGGMLSKTDSKDGAKSHKDLIEQLIAESKKRKAEKQKDKEQTLDLTEKLDSEWKDLQPVVFKRPRVEEESAIDKVLNKAEKNSQDYDKMMRELKFEKRGTPSDALKSDEKQAKEEREKLEQWERERQQRMTGDDEVIPVRVKQPAPKHRSAEDLDDNFELEDEASFMLAYDKDGKPLVPEHILKDYSVPNAKPKFEDGVSDTSDEENSDDADDESNGDDDGDSDEQEVSDEDAPEETTDDEKHVNDTDNKGAAKEINDNRVEGKKLEVENLSSQSESEEDTEDELNVNIEEGTKVKEPQKVTTQIQINNVDTSEDSSNDIPESNDKQNVLKITEKNCISNNVSAKCNGNVINENNRKNVDDLNAFNDLKSSSDESDDSDVDDTEIGAMFEKSNEGESLKELLSGKTPSQQYTAIQKLIKTYDPSLGEENKDKLSRLFAHLLQHINDIFTNLKDEGSVIKAFLIFDRLVPHMFDLAKLNKISTKKYLVDLLVEKRNKYMKRPKRIPDLDTLIFFKLISLLYPTSDFRHPVTTPSLLFMSEILNRSRFDDAFSISRGFFVTALILEYTVLSKRLVPSALNYLRGILYYCANTSILNQVQLVPPFMLHKMEKILNLNTDCSKMSVANKMAAKDLILESVDDDFKIRCLLISTVMLKEFFDNFNELEAQECMFEYHIKLLARIDTELYPKKVASVVSEVSQYMKECLEMKTYTPLCREKKIPKALRLYEPDVQEVFTGSKGSKLSREQAEAARLKTKLKKEMKGALREIRRDKSYIASVKLKEKMRSDNIRKEKVKQIYKEAAIQQGELNKLKRSK
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Gene 3D
CDD
ProteinModelPortal
PDB
5OQL
E-value=4.45929e-20,
Score=245
Ontologies
GO
PANTHER
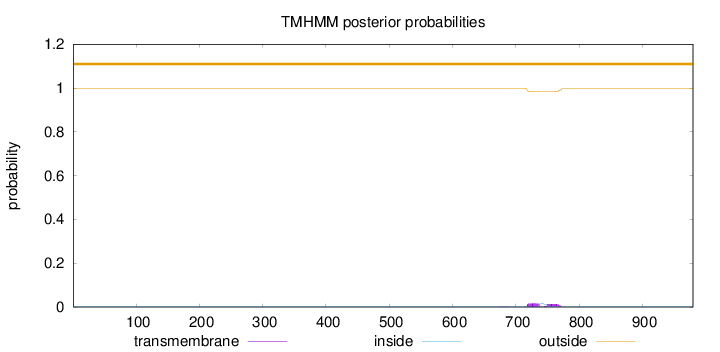
Topology
Length:
980
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.62039
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00064
outside
1 - 980
Population Genetic Test Statistics
Pi
250.419324
Theta
209.10831
Tajima's D
0.646471
CLR
0.461338
CSRT
0.549622518874056
Interpretation
Uncertain
