Pre Gene Modal
BGIBMGA010791
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_glutamyl-tRNA(Gln)_amidotransferase_subunit_A?_mitochondrial_[Bombyx_mori]
Full name
Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial
Alternative Name
Glutaminyl-tRNA synthase-like protein 1
Location in the cell
Nuclear Reliability : 2.789
Sequence
CDS
ATGAATAAATTGTCATCTTACACGATAAAAGAGATACACAGGGCGTATCGTGATAAAATACTAACGCCGACAAGTTTTGTTGATTTGTGTCTACAAAAGGCCATACAGAGTATCGAATTGAATGCCTTTACCACTATAACTGACGACGAAGCAGAAAGTCACGCAATCGAAAGCGAAAAACGACTTTACGTCGACAGAAAACCGAAGACTCTGGACGGTATTACCATCGGAATAAAAGACAATTTCTGTACTAGTAATATACCGACGACATGCGGCTCAGAAATGCTAAAAAACTTCGTTCCTACATACAACGCTACGGTATACGCAAGATTGAGGAAAGCCGGCGCTTGTTTGGTAGGAAAATGTAATTTAGACGAATTTGCAATGGGCTCCGGTACCATCGATTCCATATTCGGTCCAACCCGGAACCCATGGCAGTACGAAAGAAATAACTGGAAAATAGCTGGTGGCAGCTCTGGTGGAAGCGCTGTTGCCGTGACCACCGGATCATGCGTAGCTGCAATAGGCTCTGACACTGGTGGTTCAACTAGAAATCCAGCAGCTTTATGTGGTGTTGTTGGGTTAAAACCCTCATATGGCCTGGTGTCACGTTATGGCCTAATACCTTTAGTAAATTCAATGGATGTACCAGGAATACTGACAAGAGATATTGATGATTGTGTTGCCGTATTGAATTCAGTGGCAGGTCCTGATGACTTAGACTCTACGACTATCAAACGAGATTATAAACCAATAGAAATTCAAGATATTGATATGTCATCCGTAAAGATTGGTATACCAAAAGAATATCATTGTGAGGGGATGGATAGGGAAGTAATAGACACATGGAGTTATGTTAGTGATATTCTTGAAAAAGAAAACTGTATTGTAAAACAAGTCTCTTTGCCATATTCCTCAGCTTCAATAGTAGTATATTCTATACTGAATCAATGTGAAGTCGCAAGCAACATGGCTAGATATGATGGTATTGAATATGGTCTTAGAACTGATGAGACATATTCCACAGAAGAATTGTATGCGTCATCGAGGCTACGAGGATTCAGTGAGGTGGTAAGATCTAGGATTTTAACAGGGAACTATTTTCTACTGACGAGAAACTACAACAAATATTTTATTAAAGCTTTGAAACTACGAAGGCTTATATCAGATGATTTTAATAAAGTTTTTAATGGGGAGGATAAAGTTGATCTTCTATTGACACCAACCACATTATCTGACGCGCCGTCTTATGAAGATTTCTCATTGAAGAATAATCGTGATCAATGTGCTTTTCAAGATTTTTGCACACAACCAGCAAACATGGCCGGAATACCAGCAGTTTCAATACCGATCAGACTATCTAAAAATGGTCTTCCGTTATCCCTGCAGCTCATGGGGCCGATGTTTTCTGAAGAACTGATGTTAAATGTAGCGAAGAAGATTGAAACTAACCTTGCATCAGAAACCCTAGCGAAGCTCAACATACGCATTGACACATTACCCCAAAAATACCAACAGCTACTCGACAAACTGGCAGCAAGCCAATCATCACTTGGTATAAACCAATTAGATCACGTCTCCATATCCTTAGAGCAAACGAAAGACATTACAGAGAATCTAGAAGATGACTATGAGATTCTGAAACTTAAACACAAAGATCTAGAATTACAGAGACGGATCGAGAAGAACATAAAGTTTATCGATGAACTGAGAAGGGAATTGGAAATGTCCAGGGAGTCATTGTCTAATCAAAACCCTGGGCCAGATAATGTCCAGGATTACATAAGACAACTCAAGCAAAAGGTAGTTTCTTATGAAGAAAGCTGTACTAAAGCAAAGGTTAAATACAATAAGCTGAGCATTCCCGATACTATACTTCCCAAGTCTTTATCAGTTCTGGTGACGTCACACGAAGCCCTCCTCGAGGAGGCGAGGTCTCTCCGTCAGCGCGCCGAAGACATAACCTTCGCGAGAGACGCCATACAGAACTTGCATAGACTGAGAAGATAA
Protein
MNKLSSYTIKEIHRAYRDKILTPTSFVDLCLQKAIQSIELNAFTTITDDEAESHAIESEKRLYVDRKPKTLDGITIGIKDNFCTSNIPTTCGSEMLKNFVPTYNATVYARLRKAGACLVGKCNLDEFAMGSGTIDSIFGPTRNPWQYERNNWKIAGGSSGGSAVAVTTGSCVAAIGSDTGGSTRNPAALCGVVGLKPSYGLVSRYGLIPLVNSMDVPGILTRDIDDCVAVLNSVAGPDDLDSTTIKRDYKPIEIQDIDMSSVKIGIPKEYHCEGMDREVIDTWSYVSDILEKENCIVKQVSLPYSSASIVVYSILNQCEVASNMARYDGIEYGLRTDETYSTEELYASSRLRGFSEVVRSRILTGNYFLLTRNYNKYFIKALKLRRLISDDFNKVFNGEDKVDLLLTPTTLSDAPSYEDFSLKNNRDQCAFQDFCTQPANMAGIPAVSIPIRLSKNGLPLSLQLMGPMFSEELMLNVAKKIETNLASETLAKLNIRIDTLPQKYQQLLDKLAASQSSLGINQLDHVSISLEQTKDITENLEDDYEILKLKHKDLELQRRIEKNIKFIDELRRELEMSRESLSNQNPGPDNVQDYIRQLKQKVVSYEESCTKAKVKYNKLSIPDTILPKSLSVLVTSHEALLEEARSLRQRAEDITFARDAIQNLHRLRR
Summary
Description
Allows the formation of correctly charged Gln-tRNA(Gln) through the transamidation of misacylated Glu-tRNA(Gln) in the mitochondria. The reaction takes place in the presence of glutamine and ATP through an activated gamma-phospho-Glu-tRNA(Gln).
Catalytic Activity
ATP + H2O + L-glutamine + L-glutamyl-tRNA(Gln) = ADP + H(+) + L-glutamate + L-glutaminyl-tRNA(Gln) + phosphate
Subunit
Subunit of the heterotrimeric GatCAB amidotransferase (AdT) complex, composed of A, B and C subunits.
Subunit of the heterotrimeric GatCAB amidotransferase (AdT) complex, composed of A (QRSL1), B (GATB) and C (GATC) subunits.
Subunit of the heterotrimeric GatCAB amidotransferase (AdT) complex, composed of A (QRSL1), B (GATB) and C (GATC) subunits.
Similarity
Belongs to the amidase family. GatA subfamily.
Keywords
ATP-binding
Complete proteome
Ligase
Mitochondrion
Nucleotide-binding
Protein biosynthesis
Reference proteome
Feature
chain Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial
Uniprot
H9JMN8
A0A2H1VJ00
A0A212EV54
A0A0L7LNI1
A0A0N1PF89
A0A194PLQ4
+ More
A0A2A4JTP0 A0A2W1BZT4 A0A3S2N5M0 A0A3B0KBW5 B3LVB4 B4MBW2 Q29BT3 B4K5Z0 B4GPP0 A0A1W4UPQ1 A0A0L0CB66 B0WAE3 B4ILV6 B4NHY9 B3P381 W8C564 B4QTW1 A0A0M3QY70 B4JYK4 H5V8A8 B4PMQ7 Q9VE09 A0A1I8MZ60 A0A1Q3F751 A0A1I8PPZ0 Q17H91 A0A2K7P689 A0A1J1IEV0 A0A182GXB9 A0A0K8UXK0 A0A182JN09 A0A182RBK1 A0A182VBM5 A0A182XFF8 A0A182FXF8 Q7QH64 A0A182I843 W5J8A2 A0A182L7R2 A0A182YF37 A0A182VX24 A0A084VTD7 A0A182T1L4 A0A182NKS4 A0A182P455 A0A182QRT8 A0A182JVJ5 D6WJE7 A0A232F8D4 K7J1T3 A0A1Y1L0R6 A0A1B0CF56 N6TK20 A0A131Z5V6 A0A224YRR3 L7M5A1 A0A1W4X127 F4WIW5 E9II25 A0A023GNM9 A0A1B6HBY6 A0A0J7NL13 A0A310SFW9 E1ZW07 A0A1B6LAF8 A0A1E1XDX6 A0A1B6FYL8 T1IY23 A0A154P122 A0A195CU11 A0A0L7RFD7 H0ZPL5 A0A0N0BBN7 A0A218V091 B7PDC5 A0A151WLH7 A0A026WY24 A0A1V4JLZ9 A0A091T0C8 T1JHV9 A0A151IXQ0 E2B9Q7 A0A0Q3UTU4
A0A2A4JTP0 A0A2W1BZT4 A0A3S2N5M0 A0A3B0KBW5 B3LVB4 B4MBW2 Q29BT3 B4K5Z0 B4GPP0 A0A1W4UPQ1 A0A0L0CB66 B0WAE3 B4ILV6 B4NHY9 B3P381 W8C564 B4QTW1 A0A0M3QY70 B4JYK4 H5V8A8 B4PMQ7 Q9VE09 A0A1I8MZ60 A0A1Q3F751 A0A1I8PPZ0 Q17H91 A0A2K7P689 A0A1J1IEV0 A0A182GXB9 A0A0K8UXK0 A0A182JN09 A0A182RBK1 A0A182VBM5 A0A182XFF8 A0A182FXF8 Q7QH64 A0A182I843 W5J8A2 A0A182L7R2 A0A182YF37 A0A182VX24 A0A084VTD7 A0A182T1L4 A0A182NKS4 A0A182P455 A0A182QRT8 A0A182JVJ5 D6WJE7 A0A232F8D4 K7J1T3 A0A1Y1L0R6 A0A1B0CF56 N6TK20 A0A131Z5V6 A0A224YRR3 L7M5A1 A0A1W4X127 F4WIW5 E9II25 A0A023GNM9 A0A1B6HBY6 A0A0J7NL13 A0A310SFW9 E1ZW07 A0A1B6LAF8 A0A1E1XDX6 A0A1B6FYL8 T1IY23 A0A154P122 A0A195CU11 A0A0L7RFD7 H0ZPL5 A0A0N0BBN7 A0A218V091 B7PDC5 A0A151WLH7 A0A026WY24 A0A1V4JLZ9 A0A091T0C8 T1JHV9 A0A151IXQ0 E2B9Q7 A0A0Q3UTU4
EC Number
6.3.5.7
Pubmed
19121390
22118469
26227816
26354079
28756777
17994087
+ More
15632085 26108605 24495485 17550304 10731132 12537572 12537569 25315136 17510324 26483478 12364791 20920257 23761445 20966253 25244985 24438588 18362917 19820115 28648823 20075255 28004739 23537049 26830274 28797301 25576852 21719571 21282665 20798317 28503490 20360741 24508170 30249741
15632085 26108605 24495485 17550304 10731132 12537572 12537569 25315136 17510324 26483478 12364791 20920257 23761445 20966253 25244985 24438588 18362917 19820115 28648823 20075255 28004739 23537049 26830274 28797301 25576852 21719571 21282665 20798317 28503490 20360741 24508170 30249741
EMBL
BABH01016560
ODYU01002556
SOQ40224.1
AGBW02012241
OWR45366.1
JTDY01000481
+ More
KOB76990.1 KQ461003 KPJ10011.1 KQ459601 KPI93664.1 NWSH01000657 PCG74984.1 KZ149908 PZC78236.1 RSAL01000369 RVE42158.1 OUUW01000007 SPP82551.1 CH902617 EDV43646.1 CH940656 EDW58583.1 CM000070 CH933806 EDW16227.1 CH479186 EDW39123.1 JRES01000657 KNC29477.1 DS231871 CH480892 EDW55432.1 CH964272 EDW83639.1 CH954181 EDV48533.1 GAMC01001947 JAC04609.1 CM000364 EDX12384.1 CP012526 ALC47163.1 CH916377 EDV90766.1 BT133209 AFA28450.1 CM000160 EDW95977.1 AE014297 AY061613 GFDL01011678 JAV23367.1 CH477252 CVRI01000048 CRK98789.1 JXUM01003476 KQ560159 KXJ84113.1 GDHF01021003 JAI31311.1 AAAB01008817 APCN01002617 ADMH02002102 ETN59100.1 ATLV01016349 KE525079 KFB41231.1 AXCN02001191 KQ971342 EFA03686.1 NNAY01000698 OXU26925.1 GEZM01070640 JAV66371.1 AJWK01009713 APGK01021752 APGK01021753 APGK01021754 KB740358 KB631949 ENN80814.1 ERL87418.1 GEDV01002385 JAP86172.1 GFPF01006135 MAA17281.1 GACK01006765 JAA58269.1 GL888177 EGI65911.1 GL763347 EFZ19768.1 GBBM01000893 JAC34525.1 GECU01035482 JAS72224.1 LBMM01003786 KMQ93135.1 KQ768478 OAD53139.1 GL434713 EFN74629.1 GEBQ01019294 JAT20683.1 GFAC01001820 JAT97368.1 GECZ01014491 JAS55278.1 JH431669 KQ434796 KZC05626.1 KQ977279 KYN04178.1 KQ414608 KOC69436.1 ABQF01000466 KQ435970 KOX67811.1 MUZQ01000087 OWK59032.1 DS689337 KQ982959 KYQ48670.1 KK107077 QOIP01000001 EZA60616.1 RLU27107.1 LSYS01006902 OPJ73226.1 KK494304 KFQ64747.1 JH431998 KQ980796 KYN12972.1 GL446584 EFN87591.1 LMAW01000836 KQK85274.1
KOB76990.1 KQ461003 KPJ10011.1 KQ459601 KPI93664.1 NWSH01000657 PCG74984.1 KZ149908 PZC78236.1 RSAL01000369 RVE42158.1 OUUW01000007 SPP82551.1 CH902617 EDV43646.1 CH940656 EDW58583.1 CM000070 CH933806 EDW16227.1 CH479186 EDW39123.1 JRES01000657 KNC29477.1 DS231871 CH480892 EDW55432.1 CH964272 EDW83639.1 CH954181 EDV48533.1 GAMC01001947 JAC04609.1 CM000364 EDX12384.1 CP012526 ALC47163.1 CH916377 EDV90766.1 BT133209 AFA28450.1 CM000160 EDW95977.1 AE014297 AY061613 GFDL01011678 JAV23367.1 CH477252 CVRI01000048 CRK98789.1 JXUM01003476 KQ560159 KXJ84113.1 GDHF01021003 JAI31311.1 AAAB01008817 APCN01002617 ADMH02002102 ETN59100.1 ATLV01016349 KE525079 KFB41231.1 AXCN02001191 KQ971342 EFA03686.1 NNAY01000698 OXU26925.1 GEZM01070640 JAV66371.1 AJWK01009713 APGK01021752 APGK01021753 APGK01021754 KB740358 KB631949 ENN80814.1 ERL87418.1 GEDV01002385 JAP86172.1 GFPF01006135 MAA17281.1 GACK01006765 JAA58269.1 GL888177 EGI65911.1 GL763347 EFZ19768.1 GBBM01000893 JAC34525.1 GECU01035482 JAS72224.1 LBMM01003786 KMQ93135.1 KQ768478 OAD53139.1 GL434713 EFN74629.1 GEBQ01019294 JAT20683.1 GFAC01001820 JAT97368.1 GECZ01014491 JAS55278.1 JH431669 KQ434796 KZC05626.1 KQ977279 KYN04178.1 KQ414608 KOC69436.1 ABQF01000466 KQ435970 KOX67811.1 MUZQ01000087 OWK59032.1 DS689337 KQ982959 KYQ48670.1 KK107077 QOIP01000001 EZA60616.1 RLU27107.1 LSYS01006902 OPJ73226.1 KK494304 KFQ64747.1 JH431998 KQ980796 KYN12972.1 GL446584 EFN87591.1 LMAW01000836 KQK85274.1
Proteomes
UP000005204
UP000007151
UP000037510
UP000053240
UP000053268
UP000218220
+ More
UP000283053 UP000268350 UP000007801 UP000008792 UP000001819 UP000009192 UP000008744 UP000192221 UP000037069 UP000002320 UP000001292 UP000007798 UP000008711 UP000000304 UP000092553 UP000001070 UP000002282 UP000000803 UP000095301 UP000095300 UP000008820 UP000183832 UP000069940 UP000249989 UP000075880 UP000075900 UP000075903 UP000076407 UP000069272 UP000007062 UP000075840 UP000000673 UP000075882 UP000076408 UP000075920 UP000030765 UP000075901 UP000075884 UP000075885 UP000075886 UP000075881 UP000007266 UP000215335 UP000002358 UP000092461 UP000019118 UP000030742 UP000192223 UP000007755 UP000036403 UP000000311 UP000076502 UP000078542 UP000053825 UP000007754 UP000053105 UP000197619 UP000001555 UP000075809 UP000053097 UP000279307 UP000190648 UP000078492 UP000008237 UP000051836
UP000283053 UP000268350 UP000007801 UP000008792 UP000001819 UP000009192 UP000008744 UP000192221 UP000037069 UP000002320 UP000001292 UP000007798 UP000008711 UP000000304 UP000092553 UP000001070 UP000002282 UP000000803 UP000095301 UP000095300 UP000008820 UP000183832 UP000069940 UP000249989 UP000075880 UP000075900 UP000075903 UP000076407 UP000069272 UP000007062 UP000075840 UP000000673 UP000075882 UP000076408 UP000075920 UP000030765 UP000075901 UP000075884 UP000075885 UP000075886 UP000075881 UP000007266 UP000215335 UP000002358 UP000092461 UP000019118 UP000030742 UP000192223 UP000007755 UP000036403 UP000000311 UP000076502 UP000078542 UP000053825 UP000007754 UP000053105 UP000197619 UP000001555 UP000075809 UP000053097 UP000279307 UP000190648 UP000078492 UP000008237 UP000051836
Pfam
PF01425 Amidase
SUPFAM
SSF75304
SSF75304
Gene 3D
ProteinModelPortal
H9JMN8
A0A2H1VJ00
A0A212EV54
A0A0L7LNI1
A0A0N1PF89
A0A194PLQ4
+ More
A0A2A4JTP0 A0A2W1BZT4 A0A3S2N5M0 A0A3B0KBW5 B3LVB4 B4MBW2 Q29BT3 B4K5Z0 B4GPP0 A0A1W4UPQ1 A0A0L0CB66 B0WAE3 B4ILV6 B4NHY9 B3P381 W8C564 B4QTW1 A0A0M3QY70 B4JYK4 H5V8A8 B4PMQ7 Q9VE09 A0A1I8MZ60 A0A1Q3F751 A0A1I8PPZ0 Q17H91 A0A2K7P689 A0A1J1IEV0 A0A182GXB9 A0A0K8UXK0 A0A182JN09 A0A182RBK1 A0A182VBM5 A0A182XFF8 A0A182FXF8 Q7QH64 A0A182I843 W5J8A2 A0A182L7R2 A0A182YF37 A0A182VX24 A0A084VTD7 A0A182T1L4 A0A182NKS4 A0A182P455 A0A182QRT8 A0A182JVJ5 D6WJE7 A0A232F8D4 K7J1T3 A0A1Y1L0R6 A0A1B0CF56 N6TK20 A0A131Z5V6 A0A224YRR3 L7M5A1 A0A1W4X127 F4WIW5 E9II25 A0A023GNM9 A0A1B6HBY6 A0A0J7NL13 A0A310SFW9 E1ZW07 A0A1B6LAF8 A0A1E1XDX6 A0A1B6FYL8 T1IY23 A0A154P122 A0A195CU11 A0A0L7RFD7 H0ZPL5 A0A0N0BBN7 A0A218V091 B7PDC5 A0A151WLH7 A0A026WY24 A0A1V4JLZ9 A0A091T0C8 T1JHV9 A0A151IXQ0 E2B9Q7 A0A0Q3UTU4
A0A2A4JTP0 A0A2W1BZT4 A0A3S2N5M0 A0A3B0KBW5 B3LVB4 B4MBW2 Q29BT3 B4K5Z0 B4GPP0 A0A1W4UPQ1 A0A0L0CB66 B0WAE3 B4ILV6 B4NHY9 B3P381 W8C564 B4QTW1 A0A0M3QY70 B4JYK4 H5V8A8 B4PMQ7 Q9VE09 A0A1I8MZ60 A0A1Q3F751 A0A1I8PPZ0 Q17H91 A0A2K7P689 A0A1J1IEV0 A0A182GXB9 A0A0K8UXK0 A0A182JN09 A0A182RBK1 A0A182VBM5 A0A182XFF8 A0A182FXF8 Q7QH64 A0A182I843 W5J8A2 A0A182L7R2 A0A182YF37 A0A182VX24 A0A084VTD7 A0A182T1L4 A0A182NKS4 A0A182P455 A0A182QRT8 A0A182JVJ5 D6WJE7 A0A232F8D4 K7J1T3 A0A1Y1L0R6 A0A1B0CF56 N6TK20 A0A131Z5V6 A0A224YRR3 L7M5A1 A0A1W4X127 F4WIW5 E9II25 A0A023GNM9 A0A1B6HBY6 A0A0J7NL13 A0A310SFW9 E1ZW07 A0A1B6LAF8 A0A1E1XDX6 A0A1B6FYL8 T1IY23 A0A154P122 A0A195CU11 A0A0L7RFD7 H0ZPL5 A0A0N0BBN7 A0A218V091 B7PDC5 A0A151WLH7 A0A026WY24 A0A1V4JLZ9 A0A091T0C8 T1JHV9 A0A151IXQ0 E2B9Q7 A0A0Q3UTU4
PDB
4WJ3
E-value=1.59168e-82,
Score=782
Ontologies
PATHWAY
GO
PANTHER
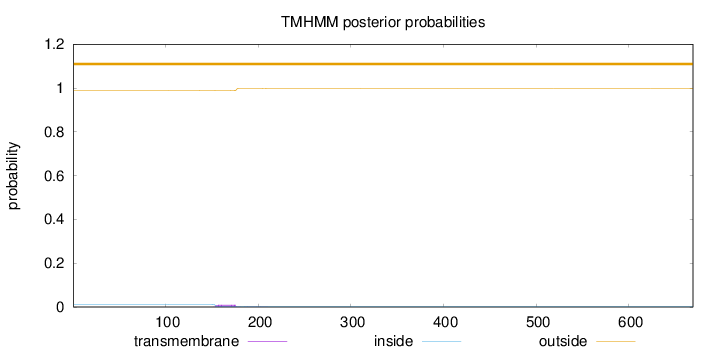
Topology
Subcellular location
Mitochondrion
Length:
669
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24762
Exp number, first 60 AAs:
0.00124
Total prob of N-in:
0.01212
outside
1 - 669
Population Genetic Test Statistics
Pi
235.837643
Theta
189.747976
Tajima's D
0.686397
CLR
15.530454
CSRT
0.569821508924554
Interpretation
Uncertain
