Pre Gene Modal
BGIBMGA010761
Annotation
PREDICTED:_ABC_transporter_G_family_member_23_isoform_X2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.706
Sequence
CDS
ATGACCGTCGCTAAAGGCACCATATACGGTCTACTTGGTGCGTCGGGCTGTGGGAAGACCACGCTTCTGTCTTGCATCGTGGGCAGACGGAAATTAAACAGCGGCGAGATATGGGTGCTCGGAGGGAAGCCTGGAACAAAAGGGTCTGGTGTCCCCGGGAAACGCGTCGGCTACATGCCTCAGGAGATAGCTCTGTACGGAGAGTTCACCATCAAGGAGACCATGATGTATTTCGGCTGGATCTTCGGCATGGAGACGAAGGAGATCGTGGATCGTCTTCGGTTCCTGCTAGACTTTCTGGACCTGCCCAGCGAGAACCGAATGGTCAAGAACTTGAGTGGAGGGCAGCAGAGAAGAGTCTCGTTCGCTGTAGCGCTGATGCACGACCCCGAACTCCTGATCTTGGACGAGCCGACGGTCGGAGTGGATCCACTGCTGAGGCAGTCGATCTGGACCCATTTGGTTCGGATCACCAGCTCCGGAGACAAGACCGTTATTATCACCACTCATTATATTGAGGAGGCCAGGCAGGCGCACTGCATTGGTCTGATGAGGAGCGGTCGTCTCCTCGCCGAGGAGTCCCCACAGGCCTTACTTAGAATGTACAGCTGCATATCGTTGGAAGATGTCTTCCTAAAACTATCCAGAAAACAGGGTCAATCGAATCAAGTAGTGGAACTAAACGTGACCGGCGGAGGAGCCCTCGGTCTGAACAAGATGTGTAAGCGCGAGGAGGCGCCCTACGCGGCCGAGGACGGGCAGGTCGTGGGCCTGAACTTCCACCAGTCCAAGGAGGTGCTCATTGTAGACCACGGAGCTGGGTCTAACGGAGACATTCCCGGTAAAGTCACGGAGGTCGTGGCCGCGGAGTGCGAAGACTGCGGGAATTGTTTCAATCTGACGTCGAGAGGGAAGATAAAAGCGTTGATCCAGAAGAACTTCCTGCGCATGTGGAGGAACATCGGCGTGATGCTGTTCATCTTCGTTCTCCCAGTGATGCAAGTCATTCTCTTCTGTCTGGCCATCGGAAGGGATCCGACAGGACTACGGATCGCCGTAGTCAACGAAGACGTGAGGATAATAGACGGCGACTGTCCCTACAACCACACCTGCTCAATGAAGAACCTCTCCTGCCGATACCTGGACAACCTGAGCGACCACATGATAAAGGAGTACTACGCTGATCTGTCGTCAGCGCTGGATGCCGTCCGTCAGGGTGAAGCCTGGGGCGCTCTGTACTTCAATGAGAATTATACGGATTCTTTGGTCGCCAGATTGGCTCTGGCTGACACGGCCGACGAAGAGACGGTGACATCGTCCGAGGTTCAAGTCTGGCTAGACATGTCCAACCAGCAGATCGGACTTATGCTCAACCGGGACATACAGTTCTCGTACAGGGACTTCGCTAAGGGTCTGCTTGAGACGTGCGACTACAACCCGAAGCTGGGCGACATCCCGATCGACTTCATGGACCCCATATACGGGAATAAGAACCCTTCGTTCACAGACTTCGTAGCTCCCGGCGTCATATTAACCATCGTATTCTTCCTCGCCGTGGCCCTGACATCGTCAGCGTTGATCGTGGAACGGATGGAGGGCCTCCTGGACCGATCCTGGGTGGCAGGGGTGTCTCCCGGAGAGATTCTGTTCTCGCACGTCGTCACCCAGTTCGTGGTGATGTGCGGGCAGACGGCGCTGGTGCTCATCTTCATGATATCGGTGTTCGGCGTCAAGAATAATGGGAACATCGTCTTCGTTATCATGCTGACTTTGCTGCAAGGTCTGTGCGGCATGTGCTTCGGGTTCGTGATATCGGCCGCTTGCGAGCTAGAGCGGAACGCGATCCAGCTGGCCTTGGGTTCGTTCTACCCCACGCTGCTGCTGAGCGGGGTGATCTGGCCCATCGAAGGGATGCCCTGGATCCTGCGCTACGTGTCCCTCTGCCTGCCCCTCACCCTCGCCACGAACTCGCTGCGGTCCATCCTGACGCGGGGCTGGCCCTTCACGGACTCGGACGTGTACATGGGCTTCGTGTCGACGTTGGCCTGGATCGCGCTCTTCCTCATCGTAACCCTCACGATCCTGAAATTCAAACGGAATTAA
Protein
MTVAKGTIYGLLGASGCGKTTLLSCIVGRRKLNSGEIWVLGGKPGTKGSGVPGKRVGYMPQEIALYGEFTIKETMMYFGWIFGMETKEIVDRLRFLLDFLDLPSENRMVKNLSGGQQRRVSFAVALMHDPELLILDEPTVGVDPLLRQSIWTHLVRITSSGDKTVIITTHYIEEARQAHCIGLMRSGRLLAEESPQALLRMYSCISLEDVFLKLSRKQGQSNQVVELNVTGGGALGLNKMCKREEAPYAAEDGQVVGLNFHQSKEVLIVDHGAGSNGDIPGKVTEVVAAECEDCGNCFNLTSRGKIKALIQKNFLRMWRNIGVMLFIFVLPVMQVILFCLAIGRDPTGLRIAVVNEDVRIIDGDCPYNHTCSMKNLSCRYLDNLSDHMIKEYYADLSSALDAVRQGEAWGALYFNENYTDSLVARLALADTADEETVTSSEVQVWLDMSNQQIGLMLNRDIQFSYRDFAKGLLETCDYNPKLGDIPIDFMDPIYGNKNPSFTDFVAPGVILTIVFFLAVALTSSALIVERMEGLLDRSWVAGVSPGEILFSHVVTQFVVMCGQTALVLIFMISVFGVKNNGNIVFVIMLTLLQGLCGMCFGFVISAACELERNAIQLALGSFYPTLLLSGVIWPIEGMPWILRYVSLCLPLTLATNSLRSILTRGWPFTDSDVYMGFVSTLAWIALFLIVTLTILKFKRN
Summary
Uniprot
A0A2W1BZQ6
A0A2A4JA19
A0A0N1INK1
A0A212FLT5
A0A0H3VKA1
H9JMK8
+ More
A0A194PLP9 A0A023EV47 A0A1S4G863 Q17AN0 A0A084VEI8 A0A182IRQ6 A0A182P3Y2 A0A182Q700 A0A182UX41 Q7QAJ0 A0A2M4CUM8 A0A182W129 A0A182I952 A0A182M4G5 A0A182TT60 A0A2M4ARM4 A0A182WVH2 A0A182RRS9 A0A1J1IZI7 A0A182YG59 B0WAQ3 A0A1Q3FNR9 A0A1Q3FP32 A0A182FWD5 A0A232FC36 A0A1L8DYR0 K7J6H5 A0A336K4Q9 A0A1B0CMA7 E9IWQ8 A0A151ILT1 A0A182JQB6 A0A154P3U8 A0A0A9YJM1 A0A158NA05 F4WEA7 A0A151WEU6 A0A0A9Y5R6 A0A151IUM7 A0A195FI37 A0A0C9R9Y7 A0A026WIH4 A0A3L8E0Z8 A0A067RI70 E0VBP5 A0A0L7QYQ1 A0A2H8TU48 A0A1B6ILM8 A0A1B6M494 A0A310S9Q0 A0A0G3FE97 A0A1B6M8X9 E2C1L8 A0A158V0H3 J9K0F0 A0A139WBS7 A0A1Y1NJD4 A0A1Y1NFQ0 A0A1Z3FWE1 A0A139WBU4 A0A1W4WF93 A0A1B6DBU3 U5EZR5 B4K7R0 A0A0Q9X3W6 A0A0Q9XEE7 A0A0Q9WN00 B3LV93 A0A0P8YNQ3 A0A0P9CAZ6 B4M194 B4JXS8 A0A0Q9XD15 A0A0Q9X973 A0A034VF11 B4N8Q1 A0A0Q9X5B5 B4HZ08 W8AIT3 W8B731 A0A0K8VIW9 A0A3B0JEI2 A0A182KXV9 B3P5R1 A0A034VFN0 B4PQQ7 A0A1W4W2J8 A0A1W4WD52 A0A0R1E4V5 A0A1W4WEF2 I5AN16 A0A0R1E588 A0A0M4EG09 W5JDV2
A0A194PLP9 A0A023EV47 A0A1S4G863 Q17AN0 A0A084VEI8 A0A182IRQ6 A0A182P3Y2 A0A182Q700 A0A182UX41 Q7QAJ0 A0A2M4CUM8 A0A182W129 A0A182I952 A0A182M4G5 A0A182TT60 A0A2M4ARM4 A0A182WVH2 A0A182RRS9 A0A1J1IZI7 A0A182YG59 B0WAQ3 A0A1Q3FNR9 A0A1Q3FP32 A0A182FWD5 A0A232FC36 A0A1L8DYR0 K7J6H5 A0A336K4Q9 A0A1B0CMA7 E9IWQ8 A0A151ILT1 A0A182JQB6 A0A154P3U8 A0A0A9YJM1 A0A158NA05 F4WEA7 A0A151WEU6 A0A0A9Y5R6 A0A151IUM7 A0A195FI37 A0A0C9R9Y7 A0A026WIH4 A0A3L8E0Z8 A0A067RI70 E0VBP5 A0A0L7QYQ1 A0A2H8TU48 A0A1B6ILM8 A0A1B6M494 A0A310S9Q0 A0A0G3FE97 A0A1B6M8X9 E2C1L8 A0A158V0H3 J9K0F0 A0A139WBS7 A0A1Y1NJD4 A0A1Y1NFQ0 A0A1Z3FWE1 A0A139WBU4 A0A1W4WF93 A0A1B6DBU3 U5EZR5 B4K7R0 A0A0Q9X3W6 A0A0Q9XEE7 A0A0Q9WN00 B3LV93 A0A0P8YNQ3 A0A0P9CAZ6 B4M194 B4JXS8 A0A0Q9XD15 A0A0Q9X973 A0A034VF11 B4N8Q1 A0A0Q9X5B5 B4HZ08 W8AIT3 W8B731 A0A0K8VIW9 A0A3B0JEI2 A0A182KXV9 B3P5R1 A0A034VFN0 B4PQQ7 A0A1W4W2J8 A0A1W4WD52 A0A0R1E4V5 A0A1W4WEF2 I5AN16 A0A0R1E588 A0A0M4EG09 W5JDV2
Pubmed
28756777
26354079
22118469
26333918
19121390
24945155
+ More
17510324 24438588 12364791 25244985 28648823 20075255 21282665 25401762 26823975 21347285 21719571 24508170 30249741 24845553 20566863 20798317 18362917 19820115 28004739 28610908 17994087 18057021 25348373 24495485 20966253 17550304 15632085 20920257 23761445
17510324 24438588 12364791 25244985 28648823 20075255 21282665 25401762 26823975 21347285 21719571 24508170 30249741 24845553 20566863 20798317 18362917 19820115 28004739 28610908 17994087 18057021 25348373 24495485 20966253 17550304 15632085 20920257 23761445
EMBL
KZ149908
PZC78206.1
NWSH01002444
PCG68290.1
KQ461003
KPJ10017.1
+ More
AGBW02007729 OWR54659.1 KP260785 AKC96438.1 BABH01016552 BABH01016553 KQ459601 KPI93659.1 GAPW01000273 JAC13325.1 CH477332 EAT43312.1 ATLV01012278 KE524778 KFB36382.1 AXCN02001185 AAAB01008888 EAA08957.4 GGFL01004874 MBW69052.1 APCN01003191 AXCM01005332 GGFK01010093 MBW43414.1 CVRI01000064 CRL05114.1 DS231874 EDS41694.1 GFDL01005796 JAV29249.1 GFDL01005797 JAV29248.1 NNAY01000487 OXU28050.1 GFDF01002652 JAV11432.1 UFQS01000026 UFQT01000026 SSW97683.1 SSX18069.1 AJWK01018516 AJWK01018517 AJWK01018518 GL766597 EFZ14974.1 KQ977104 KYN05824.1 KQ434803 KZC06014.1 GBHO01010337 GBHO01009451 GBRD01007340 GDHC01014585 JAG33267.1 JAG34153.1 JAG58481.1 JAQ04044.1 ADTU01000145 ADTU01000146 ADTU01000147 ADTU01000148 ADTU01000149 GL888102 EGI67545.1 KQ983238 KYQ46326.1 GBHO01016100 GBHO01010341 JAG27504.1 JAG33263.1 KQ980956 KYN11092.1 KQ981606 KYN39654.1 GBYB01005060 GBYB01005062 GBYB01005063 GBYB01005147 JAG74827.1 JAG74829.1 JAG74830.1 JAG74914.1 KK107203 EZA55461.1 QOIP01000002 RLU25698.1 KK852498 KDR22698.1 DS235035 EEB10801.1 KQ414688 KOC63687.1 GFXV01005297 MBW17102.1 GECU01035592 GECU01019890 JAS72114.1 JAS87816.1 GEBQ01009237 JAT30740.1 KQ767877 OAD53261.1 KP343867 AKJ85499.1 GEBQ01007609 JAT32368.1 GL451937 EFN78194.1 KF828806 AIN44123.1 ABLF02035201 ABLF02035202 KQ971372 KYB25372.1 GEZM01003527 JAV96755.1 GEZM01003528 JAV96754.1 KY792990 ASC49547.1 KYB25373.1 GEDC01014139 JAS23159.1 GANO01000296 JAB59575.1 CH933806 EDW16431.1 KRG02121.1 KRG02123.1 KRG02120.1 CH940650 KRF83319.1 CH902617 EDV43625.1 KPU80390.1 KPU80391.1 EDW67505.1 CH916377 EDV90490.1 KRG02119.1 KRG02122.1 GAKP01018587 JAC40365.1 CH964232 EDW81502.1 KRF99463.1 CH480819 EDW53265.1 GAMC01020938 JAB85617.1 GAMC01020936 JAB85619.1 GDHF01018144 GDHF01013480 JAI34170.1 JAI38834.1 OUUW01000005 SPP80794.1 CH954182 EDV53311.1 GAKP01018589 JAC40363.1 CM000160 EDW98396.1 KRK04251.1 CM000070 EIM52351.2 KRK04249.1 CP012526 ALC47216.1 ADMH02001506 ETN62261.1
AGBW02007729 OWR54659.1 KP260785 AKC96438.1 BABH01016552 BABH01016553 KQ459601 KPI93659.1 GAPW01000273 JAC13325.1 CH477332 EAT43312.1 ATLV01012278 KE524778 KFB36382.1 AXCN02001185 AAAB01008888 EAA08957.4 GGFL01004874 MBW69052.1 APCN01003191 AXCM01005332 GGFK01010093 MBW43414.1 CVRI01000064 CRL05114.1 DS231874 EDS41694.1 GFDL01005796 JAV29249.1 GFDL01005797 JAV29248.1 NNAY01000487 OXU28050.1 GFDF01002652 JAV11432.1 UFQS01000026 UFQT01000026 SSW97683.1 SSX18069.1 AJWK01018516 AJWK01018517 AJWK01018518 GL766597 EFZ14974.1 KQ977104 KYN05824.1 KQ434803 KZC06014.1 GBHO01010337 GBHO01009451 GBRD01007340 GDHC01014585 JAG33267.1 JAG34153.1 JAG58481.1 JAQ04044.1 ADTU01000145 ADTU01000146 ADTU01000147 ADTU01000148 ADTU01000149 GL888102 EGI67545.1 KQ983238 KYQ46326.1 GBHO01016100 GBHO01010341 JAG27504.1 JAG33263.1 KQ980956 KYN11092.1 KQ981606 KYN39654.1 GBYB01005060 GBYB01005062 GBYB01005063 GBYB01005147 JAG74827.1 JAG74829.1 JAG74830.1 JAG74914.1 KK107203 EZA55461.1 QOIP01000002 RLU25698.1 KK852498 KDR22698.1 DS235035 EEB10801.1 KQ414688 KOC63687.1 GFXV01005297 MBW17102.1 GECU01035592 GECU01019890 JAS72114.1 JAS87816.1 GEBQ01009237 JAT30740.1 KQ767877 OAD53261.1 KP343867 AKJ85499.1 GEBQ01007609 JAT32368.1 GL451937 EFN78194.1 KF828806 AIN44123.1 ABLF02035201 ABLF02035202 KQ971372 KYB25372.1 GEZM01003527 JAV96755.1 GEZM01003528 JAV96754.1 KY792990 ASC49547.1 KYB25373.1 GEDC01014139 JAS23159.1 GANO01000296 JAB59575.1 CH933806 EDW16431.1 KRG02121.1 KRG02123.1 KRG02120.1 CH940650 KRF83319.1 CH902617 EDV43625.1 KPU80390.1 KPU80391.1 EDW67505.1 CH916377 EDV90490.1 KRG02119.1 KRG02122.1 GAKP01018587 JAC40365.1 CH964232 EDW81502.1 KRF99463.1 CH480819 EDW53265.1 GAMC01020938 JAB85617.1 GAMC01020936 JAB85619.1 GDHF01018144 GDHF01013480 JAI34170.1 JAI38834.1 OUUW01000005 SPP80794.1 CH954182 EDV53311.1 GAKP01018589 JAC40363.1 CM000160 EDW98396.1 KRK04251.1 CM000070 EIM52351.2 KRK04249.1 CP012526 ALC47216.1 ADMH02001506 ETN62261.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000005204
UP000053268
UP000008820
+ More
UP000030765 UP000075880 UP000075885 UP000075886 UP000075903 UP000007062 UP000075920 UP000075840 UP000075883 UP000075902 UP000076407 UP000075900 UP000183832 UP000076408 UP000002320 UP000069272 UP000215335 UP000002358 UP000092461 UP000078542 UP000075881 UP000076502 UP000005205 UP000007755 UP000075809 UP000078492 UP000078541 UP000053097 UP000279307 UP000027135 UP000009046 UP000053825 UP000008237 UP000007819 UP000007266 UP000192223 UP000009192 UP000008792 UP000007801 UP000001070 UP000007798 UP000001292 UP000268350 UP000075882 UP000008711 UP000002282 UP000192221 UP000001819 UP000092553 UP000000673
UP000030765 UP000075880 UP000075885 UP000075886 UP000075903 UP000007062 UP000075920 UP000075840 UP000075883 UP000075902 UP000076407 UP000075900 UP000183832 UP000076408 UP000002320 UP000069272 UP000215335 UP000002358 UP000092461 UP000078542 UP000075881 UP000076502 UP000005205 UP000007755 UP000075809 UP000078492 UP000078541 UP000053097 UP000279307 UP000027135 UP000009046 UP000053825 UP000008237 UP000007819 UP000007266 UP000192223 UP000009192 UP000008792 UP000007801 UP000001070 UP000007798 UP000001292 UP000268350 UP000075882 UP000008711 UP000002282 UP000192221 UP000001819 UP000092553 UP000000673
PRIDE
Pfam
Interpro
IPR013525
ABC_2_trans
+ More
IPR003593 AAA+_ATPase
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR000412 ABC_2_transport
IPR011626 Alpha-macroglobulin_TED
IPR036595 A-macroglobulin_rcpt-bd_sf
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR041555 MG3
IPR013783 Ig-like_fold
IPR019742 MacrogloblnA2_CS
IPR002890 MG2
IPR041813 A2M_TED
IPR011625 A2M_N_BRD
IPR001599 Macroglobln_a2
IPR009048 A-macroglobulin_rcpt-bd
IPR016024 ARM-type_fold
IPR028426 Huntingtin_fam
IPR003593 AAA+_ATPase
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR000412 ABC_2_transport
IPR011626 Alpha-macroglobulin_TED
IPR036595 A-macroglobulin_rcpt-bd_sf
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR041555 MG3
IPR013783 Ig-like_fold
IPR019742 MacrogloblnA2_CS
IPR002890 MG2
IPR041813 A2M_TED
IPR011625 A2M_N_BRD
IPR001599 Macroglobln_a2
IPR009048 A-macroglobulin_rcpt-bd
IPR016024 ARM-type_fold
IPR028426 Huntingtin_fam
Gene 3D
CDD
ProteinModelPortal
A0A2W1BZQ6
A0A2A4JA19
A0A0N1INK1
A0A212FLT5
A0A0H3VKA1
H9JMK8
+ More
A0A194PLP9 A0A023EV47 A0A1S4G863 Q17AN0 A0A084VEI8 A0A182IRQ6 A0A182P3Y2 A0A182Q700 A0A182UX41 Q7QAJ0 A0A2M4CUM8 A0A182W129 A0A182I952 A0A182M4G5 A0A182TT60 A0A2M4ARM4 A0A182WVH2 A0A182RRS9 A0A1J1IZI7 A0A182YG59 B0WAQ3 A0A1Q3FNR9 A0A1Q3FP32 A0A182FWD5 A0A232FC36 A0A1L8DYR0 K7J6H5 A0A336K4Q9 A0A1B0CMA7 E9IWQ8 A0A151ILT1 A0A182JQB6 A0A154P3U8 A0A0A9YJM1 A0A158NA05 F4WEA7 A0A151WEU6 A0A0A9Y5R6 A0A151IUM7 A0A195FI37 A0A0C9R9Y7 A0A026WIH4 A0A3L8E0Z8 A0A067RI70 E0VBP5 A0A0L7QYQ1 A0A2H8TU48 A0A1B6ILM8 A0A1B6M494 A0A310S9Q0 A0A0G3FE97 A0A1B6M8X9 E2C1L8 A0A158V0H3 J9K0F0 A0A139WBS7 A0A1Y1NJD4 A0A1Y1NFQ0 A0A1Z3FWE1 A0A139WBU4 A0A1W4WF93 A0A1B6DBU3 U5EZR5 B4K7R0 A0A0Q9X3W6 A0A0Q9XEE7 A0A0Q9WN00 B3LV93 A0A0P8YNQ3 A0A0P9CAZ6 B4M194 B4JXS8 A0A0Q9XD15 A0A0Q9X973 A0A034VF11 B4N8Q1 A0A0Q9X5B5 B4HZ08 W8AIT3 W8B731 A0A0K8VIW9 A0A3B0JEI2 A0A182KXV9 B3P5R1 A0A034VFN0 B4PQQ7 A0A1W4W2J8 A0A1W4WD52 A0A0R1E4V5 A0A1W4WEF2 I5AN16 A0A0R1E588 A0A0M4EG09 W5JDV2
A0A194PLP9 A0A023EV47 A0A1S4G863 Q17AN0 A0A084VEI8 A0A182IRQ6 A0A182P3Y2 A0A182Q700 A0A182UX41 Q7QAJ0 A0A2M4CUM8 A0A182W129 A0A182I952 A0A182M4G5 A0A182TT60 A0A2M4ARM4 A0A182WVH2 A0A182RRS9 A0A1J1IZI7 A0A182YG59 B0WAQ3 A0A1Q3FNR9 A0A1Q3FP32 A0A182FWD5 A0A232FC36 A0A1L8DYR0 K7J6H5 A0A336K4Q9 A0A1B0CMA7 E9IWQ8 A0A151ILT1 A0A182JQB6 A0A154P3U8 A0A0A9YJM1 A0A158NA05 F4WEA7 A0A151WEU6 A0A0A9Y5R6 A0A151IUM7 A0A195FI37 A0A0C9R9Y7 A0A026WIH4 A0A3L8E0Z8 A0A067RI70 E0VBP5 A0A0L7QYQ1 A0A2H8TU48 A0A1B6ILM8 A0A1B6M494 A0A310S9Q0 A0A0G3FE97 A0A1B6M8X9 E2C1L8 A0A158V0H3 J9K0F0 A0A139WBS7 A0A1Y1NJD4 A0A1Y1NFQ0 A0A1Z3FWE1 A0A139WBU4 A0A1W4WF93 A0A1B6DBU3 U5EZR5 B4K7R0 A0A0Q9X3W6 A0A0Q9XEE7 A0A0Q9WN00 B3LV93 A0A0P8YNQ3 A0A0P9CAZ6 B4M194 B4JXS8 A0A0Q9XD15 A0A0Q9X973 A0A034VF11 B4N8Q1 A0A0Q9X5B5 B4HZ08 W8AIT3 W8B731 A0A0K8VIW9 A0A3B0JEI2 A0A182KXV9 B3P5R1 A0A034VFN0 B4PQQ7 A0A1W4W2J8 A0A1W4WD52 A0A0R1E4V5 A0A1W4WEF2 I5AN16 A0A0R1E588 A0A0M4EG09 W5JDV2
PDB
4YER
E-value=2.70383e-25,
Score=289
Ontologies
GO
PANTHER
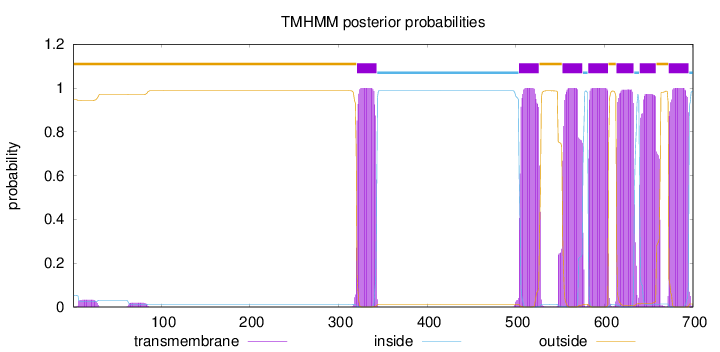
Topology
Length:
700
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
156.26475
Exp number, first 60 AAs:
0.689219999999999
Total prob of N-in:
0.05328
outside
1 - 320
TMhelix
321 - 343
inside
344 - 503
TMhelix
504 - 526
outside
527 - 552
TMhelix
553 - 575
inside
576 - 581
TMhelix
582 - 604
outside
605 - 613
TMhelix
614 - 633
inside
634 - 639
TMhelix
640 - 658
outside
659 - 672
TMhelix
673 - 695
inside
696 - 700
Population Genetic Test Statistics
Pi
213.615896
Theta
151.104674
Tajima's D
0.853309
CLR
0.256797
CSRT
0.615569221538923
Interpretation
Uncertain
