Gene
KWMTBOMO15494
Annotation
S33901_reverse_transcriptase_-_silkworm_transposon_Pao
Location in the cell
Mitochondrial Reliability : 2.227
Sequence
CDS
ATGGATGCGCTATGCATCACACCAAGAAAACCAAAAACAGACCCAGAGGAACAGGCGCTTCGCATCCTCAACAGCAATACAGTCCACACAACAGATGGAAGATACGAAACTGCTCTGCTCTGGAAAACAGATAATGTCAGTCTACCAGACAACTACAATAACTCGTTAAAGCGACTGATAAATATAGAAAACAAACTCGATCGTAATCCGGAACTGAAACAGATATACACAGAACAGATGGAATCACTCGTCACGAAAGGCTACGCCGAGCTCGCTCCAAAAACAAAAACAGAGAACAGAACGTGGTATCTACCTCACTTCGCCGTCGTGAACCCCCTGAATCCGGAAAAACTCCGAGTCGTCCACGACGCCGCCGCCAAAACAAGAGGGGTAGCTTTAAATGATATGCTGCTTAAGGGACCGGACCTACTCCAATCACTACCAGGAGTGATAATGCGATTTAGACAGCATAATATAGCAGTAACAGCAAACATCAAGGAGATGTTCATGCAAGTGAAATTGAGACACGAAGACAGAGACGCGCTCCGATATCCCTGGCGCAAAGATCGCCGAGATGACAAGCCCCCAGAAGAATACAGAATGACCTCGTTGATCTTCGGTGCGTCAAGTTCTCCTTCCACAGCAATATATGTAAAGAACTTGAACACCCAGAAACAAGAAGCCACGCACCCGGAGGCGGCAGTCGCAGTACAGAACAGACACTACGTAGACGACTTCTACTTGGATAACTTCAAAACTTTAAAAGATGCAGTGCACATAACTACAGAGTTTCATCGAAAACACGAAAGAAAGCCGACCTCGAAGACCTTCTGGACCGACAGTAAGATAGTGTTGAGATGGACAAGAACGGGATCACGCTCGAACAAACCATACGTCGCCCAACGCCTGACAGCTATAGAAGACAGTGCAACAGTAAACGGGAGGCGATGGTTACCCACGAAGCATAACTTAGCCGACGACGTGACCCGAGACGTCCCAATGCCGTACCGCAACGAACATAGGTGGTTCAGAGGGCCAGAATTCCTACGCCAACGAGAGGACCCCTGGCCGACGGAATCGGCGTCAGAAACTACAGAACCGATGGGTGAAGTAAATATAGCAGCTGCAGCACCGGCGGGAGCTTCATGGCCGAAGCGTCCCGACAACGTGGGGCGCATCGTAGACTTCAGAACCAAGGGTGGAGTTCTACGAAGACCAGTACGAAAACTACTGATCCTGCCCATCGAAGAAGACCATCCTGCACCGAAAAGAATGCGACTGACTCGCACGGCGGGAGTAATGTGCAGGACGAAATAG
Protein
MDALCITPRKPKTDPEEQALRILNSNTVHTTDGRYETALLWKTDNVSLPDNYNNSLKRLINIENKLDRNPELKQIYTEQMESLVTKGYAELAPKTKTENRTWYLPHFAVVNPLNPEKLRVVHDAAAKTRGVALNDMLLKGPDLLQSLPGVIMRFRQHNIAVTANIKEMFMQVKLRHEDRDALRYPWRKDRRDDKPPEEYRMTSLIFGASSSPSTAIYVKNLNTQKQEATHPEAAVAVQNRHYVDDFYLDNFKTLKDAVHITTEFHRKHERKPTSKTFWTDSKIVLRWTRTGSRSNKPYVAQRLTAIEDSATVNGRRWLPTKHNLADDVTRDVPMPYRNEHRWFRGPEFLRQREDPWPTESASETTEPMGEVNIAAAAPAGASWPKRPDNVGRIVDFRTKGGVLRRPVRKLLILPIEEDHPAPKRMRLTRTAGVMCRTK
Summary
Uniprot
EMBL
L09635
AAA75005.1
GGFK01002631
MBW35952.1
GGFK01002619
MBW35940.1
+ More
GGFK01002641 MBW35962.1 GGFK01002635 MBW35956.1 GGFK01002609 MBW35930.1 AAZX01006146 JXUM01136032 KQ568367 KXJ69026.1 GEHC01000967 JAV46678.1 KQ459986 KPJ18861.1 GGFM01007024 MBW27775.1 GGFL01005193 MBW69371.1 GGFK01009497 MBW42818.1 JXUM01109099 KQ565290 KXJ71123.1 GAMC01017930 JAB88625.1 GEHC01000872 JAV46773.1 GDUN01000090 JAN95829.1 JXUM01108242 KQ565213 KXJ71211.1 GGFK01008809 MBW42130.1 JXUM01034199 KQ561015 KXJ80028.1
GGFK01002641 MBW35962.1 GGFK01002635 MBW35956.1 GGFK01002609 MBW35930.1 AAZX01006146 JXUM01136032 KQ568367 KXJ69026.1 GEHC01000967 JAV46678.1 KQ459986 KPJ18861.1 GGFM01007024 MBW27775.1 GGFL01005193 MBW69371.1 GGFK01009497 MBW42818.1 JXUM01109099 KQ565290 KXJ71123.1 GAMC01017930 JAB88625.1 GEHC01000872 JAV46773.1 GDUN01000090 JAN95829.1 JXUM01108242 KQ565213 KXJ71211.1 GGFK01008809 MBW42130.1 JXUM01034199 KQ561015 KXJ80028.1
Proteomes
PRIDE
Pfam
Interpro
IPR005312
DUF1759
+ More
IPR001878 Znf_CCHC
IPR008042 Retrotrans_Pao
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR000253 FHA_dom
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR008042 Retrotrans_Pao
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR000253 FHA_dom
IPR000477 RT_dom
IPR036875 Znf_CCHC_sf
Gene 3D
ProteinModelPortal
Ontologies
KEGG
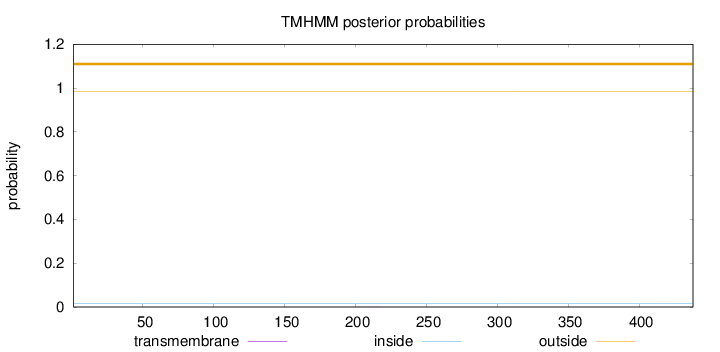
Topology
Length:
438
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0032
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01459
outside
1 - 438
Population Genetic Test Statistics
Pi
33.028939
Theta
57.056426
Tajima's D
0.730084
CLR
0.235755
CSRT
0.585370731463427
Interpretation
Uncertain
