Pre Gene Modal
BGIBMGA010764
Annotation
PREDICTED:_aminopeptidase_N_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
Mitochondrial Reliability : 1.777
Sequence
CDS
ATGGGCTCCAGTAACCACTTAACACCAGCACCTTATCGCGTCAATTATGATGCTCGTAACTGGGAACTTCTTTCGGAGACACTGCTCAGCGACAAGTACGATGAGATCCCAGTACTGGGCAGAGTGCAACTGATATCGGACGCCTTCGCTCTGGCGGCCACCAACAGACTCAACTATTCCACTACACTGATTCTTGCAAGCTACCTCCAGAGAGAGAGGGAGTACCTGCCTCTGTCGACAGCGCTGAGAGGGTTGGCCAAGATTGAGAACCTCTTGAAGCGCAGCGGAGACTACAGCGCCTTCCAGAGCTACATCAGGCATCTGGTGCAGAACTCGTATCGAAGGGCTGGAGGCCTCTCCGTCAAGCGTATCGTTAATGGCGACGATCTGAACAGCGTAAAACTGCAGGTGTTGACCAGCGGCTGGGCCTGTCGCGCGCTGATACCGGGCTGCGAGGAAAACGCCATGGAGCTGTTCCAAATGTGGATGAACACTTCCAATCCGGATGAAAATAATCCGCAAGTTTTCTTTATTTTTTCATCTAAGGACACGATCGATGGGGATCACAAGTCGATGTCTCAATTTAAAACGACCCGCGCGGGGATGAAGATCCCGCTGGACCTGCGGCGCACGGTGTGGTGCGCGGGCGCGGCGCGCGGCGGCGAGCGCGAGTGGCGCTTCCTCACGCTGCGGCTGCAGGCCGCGCGCGTTGCCGCCGCCCGCGACGCCATGCTCGCCGCCATGGCCTGCACCAGGCAGGTCTGGCTGCTCAACCAGTTCCTCGAATGGGCGGTGTCGGAGAACTCCCCGGTGCGCAAGCAGGACGCGGCCGCCGTGGTCGGTAACGTGGCCCGCACGCCGGTCGGCTACTACGTCGCGCGGGAGTTCTTCTACAACCGAATCGACGATCTGTTTAAGACTTTCAACGGTCAGAGCCGCCGTTTCGGCTATATCATCAAGACTCTGCTGGACCAGTTCACGACTCAGAGAGAATTGGACGAGTTTCTCATTTTCCACGACAAGAACCCTAAATATTTCGAGGAGACGAAGCTGGCCGTCGTTCAGGCGATTGAGAAAGCCAAAGTGAACATCGACTGGGTCGACCAAAACAAAGATTTAGTTATGAGTGAATAA
Protein
MGSSNHLTPAPYRVNYDARNWELLSETLLSDKYDEIPVLGRVQLISDAFALAATNRLNYSTTLILASYLQREREYLPLSTALRGLAKIENLLKRSGDYSAFQSYIRHLVQNSYRRAGGLSVKRIVNGDDLNSVKLQVLTSGWACRALIPGCEENAMELFQMWMNTSNPDENNPQVFFIFSSKDTIDGDHKSMSQFKTTRAGMKIPLDLRRTVWCAGAARGGEREWRFLTLRLQAARVAAARDAMLAAMACTRQVWLLNQFLEWAVSENSPVRKQDAAAVVGNVARTPVGYYVAREFFYNRIDDLFKTFNGQSRRFGYIIKTLLDQFTTQRELDEFLIFHDKNPKYFEETKLAVVQAIEKAKVNIDWVDQNKDLVMSE
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Feature
chain Aminopeptidase
Uniprot
H9JML1
I3VR83
A0A2W1BT37
A0A220K8P2
A0A0N1IFL5
A0A212EYJ4
+ More
A0A212FCL0 A0A194PRA0 A0A2A4JS21 A0A0L7L620 E0VS66 V9PBG2 D6WB37 N6UNI3 A0A336M458 U5EU86 A0A1I9WL50 A0A2L1IQ82 U4U8J3 A0A1B6HB93 A0A182ILM8 X1XT43 A0A336KKE2 A0A2S2P0U8 A0A1B6D6B8 N6TGU3 A0A1W4XNR6 A0A1Y1LUJ7 A0A182QC88 A0A084W9N5 A0A1J1IVX7 A0A182SPU1 A0A1J1IZ42 A0A182Y4U1 A0A0A1X3N6 A0A1B6M313 X1X0P5 A0A1B6LLZ7 T1DJ30 A0A2H8TQM9 W5JLT5 A0A034VBT7 A0A182FM81 A0A0K8V993 A0A0K8U897 A0A034V7H0 A0A182GQP1 A0A182VV47 Q7Q4T0 A0A182V1G0 A0A182WY28 W8BKE0 A0A182RM22 A0A023F109 A0A0N7Z8X1 A0A182K329 A0A182PL06 T1HDW0 A0A182N0H6 A0A2U9BD78 Q9VAM2 B4JYJ5 A0A182LTN9 A0A224X6V4 B4QZJ2 K7INE5 A0A069DXR4 A0A0V0G587 A0A0Q9XBS5 B4K5Y1 A0A1W4UH69 B3MTC9 A0A232EWK0 A0A310SPN7 B3MTC8 A0A1W4UIJ7 A0A1D2NEA8 Q960U1 Q29BX5 B4GPI5 A0A0L0BPV0 A0A1W4U620 Q9NH67 E2ALT1 B4PQ46 Q0KI00 B3P5I4 B4QZJ5 Q6IDH1 B4HZ87 B4QZJ3 J9JLW8 B4PQ45 B4PQ42 B3P5I7 B3P5I5 B4PQ43 A0A0R1E9Q7 A0A0R1E4R4
A0A212FCL0 A0A194PRA0 A0A2A4JS21 A0A0L7L620 E0VS66 V9PBG2 D6WB37 N6UNI3 A0A336M458 U5EU86 A0A1I9WL50 A0A2L1IQ82 U4U8J3 A0A1B6HB93 A0A182ILM8 X1XT43 A0A336KKE2 A0A2S2P0U8 A0A1B6D6B8 N6TGU3 A0A1W4XNR6 A0A1Y1LUJ7 A0A182QC88 A0A084W9N5 A0A1J1IVX7 A0A182SPU1 A0A1J1IZ42 A0A182Y4U1 A0A0A1X3N6 A0A1B6M313 X1X0P5 A0A1B6LLZ7 T1DJ30 A0A2H8TQM9 W5JLT5 A0A034VBT7 A0A182FM81 A0A0K8V993 A0A0K8U897 A0A034V7H0 A0A182GQP1 A0A182VV47 Q7Q4T0 A0A182V1G0 A0A182WY28 W8BKE0 A0A182RM22 A0A023F109 A0A0N7Z8X1 A0A182K329 A0A182PL06 T1HDW0 A0A182N0H6 A0A2U9BD78 Q9VAM2 B4JYJ5 A0A182LTN9 A0A224X6V4 B4QZJ2 K7INE5 A0A069DXR4 A0A0V0G587 A0A0Q9XBS5 B4K5Y1 A0A1W4UH69 B3MTC9 A0A232EWK0 A0A310SPN7 B3MTC8 A0A1W4UIJ7 A0A1D2NEA8 Q960U1 Q29BX5 B4GPI5 A0A0L0BPV0 A0A1W4U620 Q9NH67 E2ALT1 B4PQ46 Q0KI00 B3P5I4 B4QZJ5 Q6IDH1 B4HZ87 B4QZJ3 J9JLW8 B4PQ45 B4PQ42 B3P5I7 B3P5I5 B4PQ43 A0A0R1E9Q7 A0A0R1E4R4
EC Number
3.4.11.-
Pubmed
19121390
28756777
28630352
26354079
22118469
26227816
+ More
20566863 18362917 19820115 23537049 27538518 29415259 28004739 24438588 25244985 25830018 20920257 23761445 25348373 26483478 12364791 14747013 17210077 24495485 25474469 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 20075255 26334808 28648823 27289101 15632085 26108605 20798317 17550304 26109357 26109356
20566863 18362917 19820115 23537049 27538518 29415259 28004739 24438588 25244985 25830018 20920257 23761445 25348373 26483478 12364791 14747013 17210077 24495485 25474469 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 20075255 26334808 28648823 27289101 15632085 26108605 20798317 17550304 26109357 26109356
EMBL
BABH01016541
BABH01016542
JQ061154
AFK85028.1
KZ149908
PZC78228.1
+ More
MF319742 ASJ26456.1 KQ461003 KPJ10021.1 AGBW02011488 OWR46566.1 AGBW02009182 OWR51448.1 KQ459601 KPI93655.1 NWSH01000721 PCG74619.1 JTDY01002793 KOB70714.1 DS235745 EEB16222.1 KC282398 AGX25155.1 KQ971311 EEZ99336.1 APGK01004410 KB735736 ENN83310.1 UFQS01000417 UFQT01000417 SSX03766.1 SSX24131.1 GANO01002430 JAB57441.1 KU932234 APA33870.1 MF741655 AVD96938.1 KB632093 ERL88678.1 GECU01035837 JAS71869.1 ABLF02020107 SSX03765.1 SSX24130.1 GGMR01010385 MBY23004.1 GEDC01016064 JAS21234.1 APGK01027780 APGK01027781 APGK01027782 APGK01027783 APGK01027784 APGK01027785 KB740671 ENN79629.1 GEZM01046358 GEZM01046357 JAV77303.1 AXCN02001411 ATLV01021848 KE525324 KFB46929.1 CVRI01000060 CRL03858.1 CRL03857.1 GBXI01008343 JAD05949.1 GEBQ01009689 JAT30288.1 ABLF02020109 GEBQ01015299 JAT24678.1 GAMD01001656 JAA99934.1 GFXV01004127 MBW15932.1 ADMH02001160 ETN63860.1 GAKP01019737 JAC39215.1 GDHF01016887 JAI35427.1 GDHF01029538 JAI22776.1 GAKP01019736 JAC39216.1 JXUM01080917 KQ563216 KXJ74267.1 AAAB01008963 EAA12046.5 GAMC01009092 JAB97463.1 GBBI01003749 JAC14963.1 GDKW01002163 JAI54432.1 ACPB03023190 CP026247 AWP01958.1 AE014297 AAF56882.2 CH916377 EDV90757.1 AXCM01001470 GFTR01008270 JAW08156.1 CM000364 EDX14830.1 GBGD01000392 JAC88497.1 GECL01003570 JAP02554.1 CH933806 KRG01998.1 EDW16218.2 CH902623 EDV30519.1 NNAY01001852 OXU22728.1 KQ760397 OAD60666.1 EDV30518.1 LJIJ01000073 ODN03435.1 AY051855 AAK93279.1 CM000070 EAL26872.1 CH479186 EDW39068.1 JRES01001565 KNC22036.1 AF231040 AAF34809.1 GL440629 EFN65601.1 CM000160 EDW98315.1 BT133378 AAF56884.1 AFH36363.1 CH954182 EDV53234.1 EDX14833.1 BT014634 AAT27258.1 CH480819 EDW53344.1 EDX14831.1 EDW98314.1 EDW98311.1 EDV53237.1 EDV53235.1 EDW98312.1 KRK04210.1 KRK04208.1
MF319742 ASJ26456.1 KQ461003 KPJ10021.1 AGBW02011488 OWR46566.1 AGBW02009182 OWR51448.1 KQ459601 KPI93655.1 NWSH01000721 PCG74619.1 JTDY01002793 KOB70714.1 DS235745 EEB16222.1 KC282398 AGX25155.1 KQ971311 EEZ99336.1 APGK01004410 KB735736 ENN83310.1 UFQS01000417 UFQT01000417 SSX03766.1 SSX24131.1 GANO01002430 JAB57441.1 KU932234 APA33870.1 MF741655 AVD96938.1 KB632093 ERL88678.1 GECU01035837 JAS71869.1 ABLF02020107 SSX03765.1 SSX24130.1 GGMR01010385 MBY23004.1 GEDC01016064 JAS21234.1 APGK01027780 APGK01027781 APGK01027782 APGK01027783 APGK01027784 APGK01027785 KB740671 ENN79629.1 GEZM01046358 GEZM01046357 JAV77303.1 AXCN02001411 ATLV01021848 KE525324 KFB46929.1 CVRI01000060 CRL03858.1 CRL03857.1 GBXI01008343 JAD05949.1 GEBQ01009689 JAT30288.1 ABLF02020109 GEBQ01015299 JAT24678.1 GAMD01001656 JAA99934.1 GFXV01004127 MBW15932.1 ADMH02001160 ETN63860.1 GAKP01019737 JAC39215.1 GDHF01016887 JAI35427.1 GDHF01029538 JAI22776.1 GAKP01019736 JAC39216.1 JXUM01080917 KQ563216 KXJ74267.1 AAAB01008963 EAA12046.5 GAMC01009092 JAB97463.1 GBBI01003749 JAC14963.1 GDKW01002163 JAI54432.1 ACPB03023190 CP026247 AWP01958.1 AE014297 AAF56882.2 CH916377 EDV90757.1 AXCM01001470 GFTR01008270 JAW08156.1 CM000364 EDX14830.1 GBGD01000392 JAC88497.1 GECL01003570 JAP02554.1 CH933806 KRG01998.1 EDW16218.2 CH902623 EDV30519.1 NNAY01001852 OXU22728.1 KQ760397 OAD60666.1 EDV30518.1 LJIJ01000073 ODN03435.1 AY051855 AAK93279.1 CM000070 EAL26872.1 CH479186 EDW39068.1 JRES01001565 KNC22036.1 AF231040 AAF34809.1 GL440629 EFN65601.1 CM000160 EDW98315.1 BT133378 AAF56884.1 AFH36363.1 CH954182 EDV53234.1 EDX14833.1 BT014634 AAT27258.1 CH480819 EDW53344.1 EDX14831.1 EDW98314.1 EDW98311.1 EDV53237.1 EDV53235.1 EDW98312.1 KRK04210.1 KRK04208.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000218220
UP000037510
+ More
UP000009046 UP000007266 UP000019118 UP000030742 UP000075880 UP000007819 UP000192223 UP000075886 UP000030765 UP000183832 UP000075901 UP000076408 UP000000673 UP000069272 UP000069940 UP000249989 UP000075920 UP000007062 UP000075903 UP000076407 UP000075900 UP000075881 UP000075885 UP000015103 UP000075884 UP000246464 UP000000803 UP000001070 UP000075883 UP000000304 UP000002358 UP000009192 UP000192221 UP000007801 UP000215335 UP000094527 UP000001819 UP000008744 UP000037069 UP000000311 UP000002282 UP000008711 UP000001292
UP000009046 UP000007266 UP000019118 UP000030742 UP000075880 UP000007819 UP000192223 UP000075886 UP000030765 UP000183832 UP000075901 UP000076408 UP000000673 UP000069272 UP000069940 UP000249989 UP000075920 UP000007062 UP000075903 UP000076407 UP000075900 UP000075881 UP000075885 UP000015103 UP000075884 UP000246464 UP000000803 UP000001070 UP000075883 UP000000304 UP000002358 UP000009192 UP000192221 UP000007801 UP000215335 UP000094527 UP000001819 UP000008744 UP000037069 UP000000311 UP000002282 UP000008711 UP000001292
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JML1
I3VR83
A0A2W1BT37
A0A220K8P2
A0A0N1IFL5
A0A212EYJ4
+ More
A0A212FCL0 A0A194PRA0 A0A2A4JS21 A0A0L7L620 E0VS66 V9PBG2 D6WB37 N6UNI3 A0A336M458 U5EU86 A0A1I9WL50 A0A2L1IQ82 U4U8J3 A0A1B6HB93 A0A182ILM8 X1XT43 A0A336KKE2 A0A2S2P0U8 A0A1B6D6B8 N6TGU3 A0A1W4XNR6 A0A1Y1LUJ7 A0A182QC88 A0A084W9N5 A0A1J1IVX7 A0A182SPU1 A0A1J1IZ42 A0A182Y4U1 A0A0A1X3N6 A0A1B6M313 X1X0P5 A0A1B6LLZ7 T1DJ30 A0A2H8TQM9 W5JLT5 A0A034VBT7 A0A182FM81 A0A0K8V993 A0A0K8U897 A0A034V7H0 A0A182GQP1 A0A182VV47 Q7Q4T0 A0A182V1G0 A0A182WY28 W8BKE0 A0A182RM22 A0A023F109 A0A0N7Z8X1 A0A182K329 A0A182PL06 T1HDW0 A0A182N0H6 A0A2U9BD78 Q9VAM2 B4JYJ5 A0A182LTN9 A0A224X6V4 B4QZJ2 K7INE5 A0A069DXR4 A0A0V0G587 A0A0Q9XBS5 B4K5Y1 A0A1W4UH69 B3MTC9 A0A232EWK0 A0A310SPN7 B3MTC8 A0A1W4UIJ7 A0A1D2NEA8 Q960U1 Q29BX5 B4GPI5 A0A0L0BPV0 A0A1W4U620 Q9NH67 E2ALT1 B4PQ46 Q0KI00 B3P5I4 B4QZJ5 Q6IDH1 B4HZ87 B4QZJ3 J9JLW8 B4PQ45 B4PQ42 B3P5I7 B3P5I5 B4PQ43 A0A0R1E9Q7 A0A0R1E4R4
A0A212FCL0 A0A194PRA0 A0A2A4JS21 A0A0L7L620 E0VS66 V9PBG2 D6WB37 N6UNI3 A0A336M458 U5EU86 A0A1I9WL50 A0A2L1IQ82 U4U8J3 A0A1B6HB93 A0A182ILM8 X1XT43 A0A336KKE2 A0A2S2P0U8 A0A1B6D6B8 N6TGU3 A0A1W4XNR6 A0A1Y1LUJ7 A0A182QC88 A0A084W9N5 A0A1J1IVX7 A0A182SPU1 A0A1J1IZ42 A0A182Y4U1 A0A0A1X3N6 A0A1B6M313 X1X0P5 A0A1B6LLZ7 T1DJ30 A0A2H8TQM9 W5JLT5 A0A034VBT7 A0A182FM81 A0A0K8V993 A0A0K8U897 A0A034V7H0 A0A182GQP1 A0A182VV47 Q7Q4T0 A0A182V1G0 A0A182WY28 W8BKE0 A0A182RM22 A0A023F109 A0A0N7Z8X1 A0A182K329 A0A182PL06 T1HDW0 A0A182N0H6 A0A2U9BD78 Q9VAM2 B4JYJ5 A0A182LTN9 A0A224X6V4 B4QZJ2 K7INE5 A0A069DXR4 A0A0V0G587 A0A0Q9XBS5 B4K5Y1 A0A1W4UH69 B3MTC9 A0A232EWK0 A0A310SPN7 B3MTC8 A0A1W4UIJ7 A0A1D2NEA8 Q960U1 Q29BX5 B4GPI5 A0A0L0BPV0 A0A1W4U620 Q9NH67 E2ALT1 B4PQ46 Q0KI00 B3P5I4 B4QZJ5 Q6IDH1 B4HZ87 B4QZJ3 J9JLW8 B4PQ45 B4PQ42 B3P5I7 B3P5I5 B4PQ43 A0A0R1E9Q7 A0A0R1E4R4
PDB
5LDS
E-value=2.06968e-35,
Score=373
Ontologies
GO
PANTHER
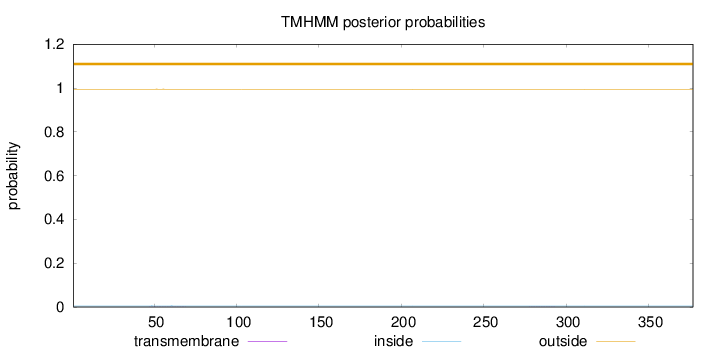
Topology
Length:
377
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01123
Exp number, first 60 AAs:
0.00477
Total prob of N-in:
0.00502
outside
1 - 377
Population Genetic Test Statistics
Pi
69.369457
Theta
139.979595
Tajima's D
-0.630956
CLR
210.486009
CSRT
0.217789110544473
Interpretation
Uncertain
