Gene
KWMTBOMO15487 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010765
Annotation
PREDICTED:_protein_Skeletor?_isoforms_B/C_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.101
Sequence
CDS
ATGTCTAAGGTTTTAGAGACTGGTTCGTTTGTTGCTATCCACGTGACGACTCCCGCCAATCTACATCCAGTGATTACGCAAGCTAAGGATAAAACCGCCCTCGGGGTTCGGCCAATGGCGCCCAAGTTGCCGCCCCTGCGCGGCCTCGCGCCTCTCTGCGTGATCCTGCTCCTGGCTGTCGGTGTAGTGACTAGCCGAAGACCAGAACCGTACTACGGAAGGCTTATAGGTCATCTAACGCAATACGCCCACGGGATTAGAGGCACAGTGTACGCAGTAGACGAGAGCACAGTCTTCATCAAGGGGTTCGCGTATGATGGGACGGGCCCTGACGCATATTTCTGGGTGGGAGATTCACCGCAGCCGTCTCCTGAAGGAACTTTAGTGCCATACCCAGAAGATTACTCGTCAAGGGATCCTCCGATATTAAGCGCTCATAGCAACACCGATATTCTGTTAAAGCTGCCAGCGGGAAAACGGTTAAGGGACATCAAATGGATCAGTGTGTGGTGTAGACGGTTTACTGTAAACTTTGGAGATGTATTCATCCCAGCCGGCTTAGATCCTCCGAGGCCAAGGGTGCTTCCGGAATTCAAGCGTCTCGCCCACGGTCTGCGGTCCGGCAACATCAGCGTCTTGGACGCTAAGACATTCTACATACCGAACTTGCACTACGACGGAGCCGGGCCTGACGCCTACTTCTGGGTCGGGAACGGATCTGAACCTAACCCTTTTGGTACCAAGGTGCCAAACGAGATGGGTTCATTGAATCCCCTCCGCGGCTACCAAGGCGAGGACATAGAGATACAGCTGCCGGCGAAGCTGACGGCCTACGACGTGGACTGGCTCGCCGTGTGGTGCGTCGAGTACAGACACAACTTCGGGCACGTTTACATCCCTAAGGACTTGGACGTGCCGCCGGCTCTGGGCCAAACTAAGATCACGACGAGTTCGACAACTTCGAAAACGCCGCACAGCGCCACCAACAACTGCAAGGAGATACTCGACAAAAGGCTGCAAGTGCGCTGGGAAATACAAGGAGACCAAGTCCAGATTACGCTATCGGCGCGTCTGCGTAAAGAGCAGTATATGGCGTTTGGCATTTCTGGAGCTGAAGGTAGACCCGCCATGCTGGGGGCGGATGTGGTTGTTGCTTATTGGGACAGCAGAGCGAACCAGCCCCGTGTGGCGGACTACACAATAACACATCTCGCTCAGTGCGACGGGGAGCGTGGGGTCTGTCCCGATCAGCGGCTGGGGGGAAGCAACGACGCAGTGCTAGTGACCGGGCGGCAACGAGACGGTGTAACGACAATAACGTACCGGCGCCCGCTGGCCGCCGCCGAGCCCTCCCGCGACAGGGAGGTGCTCTCGTCTCGGTCGCAGTCCGTCATCGCAGCCTTCGGGCCGCTCAACTCGCGCTCCGAGGCCAACGCGCACTCCTTCATGGACACCACCCGACACGACGTCCAGCTGGACTTCGGCGCGCAGAGCGACAACGCGTGCATCGGGCTGGCGGAGCTGGACGAGCTGGGCCCGGGCGCGTGGCCGGCGCGCGTGCTGTCCGGCGTCACCAACTTCACGGCGCGCATCGGGCCCGCGGGCGGCCGCCGCGGCTACACGCCCATCACTGGACATCCGTCATGGGGCATAGCGTGGTACATCAATGACCAGCTCATACCGGAGATATATGTGGAGCGCGGCAAGACTTACACCTTCTATGTCGAAGGAGGAGACGATAGAACAAATCCAGCGAGGTACCATCCGTTCTACATTAGCGATTCATCGGAAGGTGGCTTCGGACAGAAGAAGGAGGAGGAGCAGAGGAAACAGCGGGTGTTCGCGGGAGTAGCCTACGATAATGAAGGATATCCTTTCCCGACGGCCGTGGGCCGGTACTGCGAGTGGACGCACAAGTCGGTCGACCAGGCCAACGCGTCGCCCACCTTCGAGCAGTACGTGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCCAGCAGCACGCGCGCACCCTGCAGCTGGAGTGCGCGCCCGGCGACCCCGCCACGCTCGTCTGGACCGTCGCGCACGAGACGCCCGACCTGCTGTACTACCAGGTACGTTCGCTCACCGCTCCTTTACATAG
Protein
MSKVLETGSFVAIHVTTPANLHPVITQAKDKTALGVRPMAPKLPPLRGLAPLCVILLLAVGVVTSRRPEPYYGRLIGHLTQYAHGIRGTVYAVDESTVFIKGFAYDGTGPDAYFWVGDSPQPSPEGTLVPYPEDYSSRDPPILSAHSNTDILLKLPAGKRLRDIKWISVWCRRFTVNFGDVFIPAGLDPPRPRVLPEFKRLAHGLRSGNISVLDAKTFYIPNLHYDGAGPDAYFWVGNGSEPNPFGTKVPNEMGSLNPLRGYQGEDIEIQLPAKLTAYDVDWLAVWCVEYRHNFGHVYIPKDLDVPPALGQTKITTSSTTSKTPHSATNNCKEILDKRLQVRWEIQGDQVQITLSARLRKEQYMAFGISGAEGRPAMLGADVVVAYWDSRANQPRVADYTITHLAQCDGERGVCPDQRLGGSNDAVLVTGRQRDGVTTITYRRPLAAAEPSRDREVLSSRSQSVIAAFGPLNSRSEANAHSFMDTTRHDVQLDFGAQSDNACIGLAELDELGPGAWPARVLSGVTNFTARIGPAGGRRGYTPITGHPSWGIAWYINDQLIPEIYVERGKTYTFYVEGGDDRTNPARYHPFYISDSSEGGFGQKKEEEQRKQRVFAGVAYDNEGYPFPTAVGRYCEWTHKSVDQANASPTFEQYVRTLQLECAPGDPATLVWTVAHETPDLLYYQQHARTLQLECAPPPPPPPPPPPPPRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFQQHARTLQLECAPGDPATLVWTVAHETPDLLYYQVRSSSTRAPCSWSARPATPPRSSGPSRTRRPTCCTTRYVPAARAHPAAGVRARRPRHARLDRRARDARPAVLPGTFAHRSFT
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
Topology
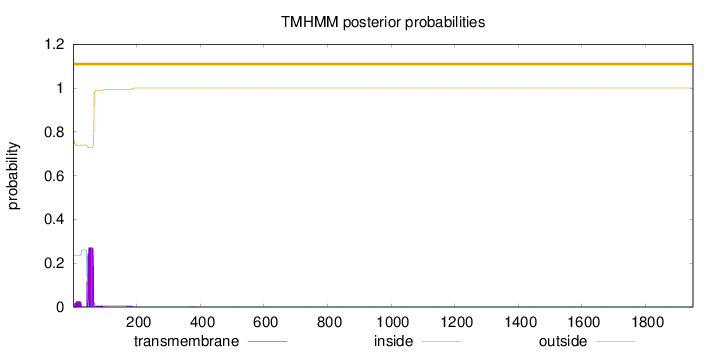
Length:
1950
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.42944
Exp number, first 60 AAs:
4.93541
Total prob of N-in:
0.23694
outside
1 - 1950
Population Genetic Test Statistics
Pi
193.281389
Theta
161.117542
Tajima's D
0.641247
CLR
0.227156
CSRT
0.555422228888556
Interpretation
Uncertain
