Gene
KWMTBOMO15484
Pre Gene Modal
BGIBMGA010784
Annotation
PREDICTED:_TBC1_domain_family_member_7-like_[Papilio_machaon]
Full name
TBC1 domain family member 7
Alternative Name
Cell migration-inducing protein 23
Location in the cell
Cytoplasmic Reliability : 3.343
Sequence
CDS
ATGACGGACGAACGTAACTTCCGTTCGTCCTACTACGAGAAAGTCGGTTGCCGCGGAGTAGAAGAGAAGAAAACATTGGAAATTCTCATGAAAGAAAAACCTTGGGATCGAGTTAAATTGAAGCAATTTTGTCTAAGATTTACAGTTTTCGCTGATTACAGGAATTTAGTCTGGAAAGTACTACTAGACATCCTGCCTGTTTACCCAGATTCGTATGAATACGTGATGAAACAGCGGACAGAGCAATATCAGGATCTCCTATATGCGGTTGAGAAGTTACAATTGATTGAACCATCAGAAATATATGACGGTTCAGGTACCCCTCAGAGCAAAGTATTCCTGGAGATGTGGGTTTTAGAAAATGAAGAATGGAAACCACCTATATTCCCAGAGACAAACTTTAATTCTGCAGATACATTTCTCCCAATTGCTAAGACACTCAGGGAACTTTATGATGATCCAGTGGATGTGTATTGGTTCAGTAAGAGATTTACAGAGGTTGTGAAGAACATGCAGAAAGATCTCAACAAATTGAAGGAAGCATTCTATAATATGTTGGACAAACAAGATGGAGAATTGTATATTTACTTGCAAGATATAAATGCATTGGAAGTAATACCATTGGTGAACTGGTTTAACTGCTGCTTTGCTGGAATATTGGATGATACATCTTTACCGAAAATATGGGACAAAGTGTGCAGCGGAGCACCTAAAATTTTAGCATATGCTGCTGTGATGCTTGTCATTACTTTACGCCGCAATATTCTAAAAGCTAAAAATGCTGATGAAGTTGTAAAATGTGTAACAGAGATTCCAAAGCAATGTGAGGAGGTAGTGGCCAACAAAGCGATTGACTTGTGGCAATATTACGGAGCCCAACCGCAAAGCGACCAACTGGCGAAGAAATGA
Protein
MTDERNFRSSYYEKVGCRGVEEKKTLEILMKEKPWDRVKLKQFCLRFTVFADYRNLVWKVLLDILPVYPDSYEYVMKQRTEQYQDLLYAVEKLQLIEPSEIYDGSGTPQSKVFLEMWVLENEEWKPPIFPETNFNSADTFLPIAKTLRELYDDPVDVYWFSKRFTEVVKNMQKDLNKLKEAFYNMLDKQDGELYIYLQDINALEVIPLVNWFNCCFAGILDDTSLPKIWDKVCSGAPKILAYAAVMLVITLRRNILKAKNADEVVKCVTEIPKQCEEVVANKAIDLWQYYGAQPQSDQLAKK
Summary
Description
Component of the TSC-TBC complex, that contains TBC1D7 in addition to the TSC1-TSC2 complex and consists of the functional complex possessing GTPase-activating protein (GAP) activity toward RHEB in response to alterations in specific cellular growth conditions. The small GTPase RHEB is a direct activator of the protein kinase activity of mTORC1 and the TSC-TBC complex acts as a negative regulator of mTORC1 signaling cascade by acting as a GAP for RHEB. Participates in the proper sensing of growth factors and glucose, but not amino acids, by mTORC1. It is unclear whether TBC1D7 acts as a GTPase-activating protein and additional studies are required to answer this question.
Subunit
Component of the TSC-TBC complex (also named Rhebulator complex), composed of the TSC1-TSC2 heterodimer and TBC1D7. Interacts with TSC1 (via C-terminal half of the coiled-coil domain).
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Keywords
Alternative splicing
Complete proteome
Cytoplasmic vesicle
GTPase activation
Reference proteome
3D-structure
Polymorphism
Feature
chain TBC1 domain family member 7
splice variant In isoform 2.
sequence variant In dbSNP:rs543580.
splice variant In isoform 2.
sequence variant In dbSNP:rs543580.
Uniprot
H9JMN1
A0A2W1BT38
A0A2A4J8V2
A0A2H1WJB3
A0A1E1WQM1
A0A0N0PBG0
+ More
S4NXH8 A0A212FCI9 A0A194PLN9 A0A0L7LCJ7 A0A2P8YJ97 A0A2J7PT35 A0A067QXG3 D6X1P7 E0VFS3 A0A336N1M4 A0A2A3EEU0 N6TM20 J3JV08 A0A232FAS6 K7IXX5 A0A1B6C0Y6 E2AMK7 A0A026W5N2 E9GLF0 U5EYJ8 U4UIC9 A0A1B6K8B6 E2BJX5 A0A182M7E1 A0A1B0GMI1 A0A1Y1KZY2 A0A0T6BG58 A0A1W4XDZ7 A0A1Q3F2R1 A0A154P214 A0A2M4AS64 A0A182HT82 A0A0L7QQT5 A0A0P5K270 A0A182YKI3 A0A1C7CZV8 Q7QC09 A0A182JX80 A0A182J6Y1 A0A182V7N6 A0A182UC53 A0A182SXF7 A0A182PA60 A0A182VX56 W5JL26 A0A0P5NLA5 A0A182XI95 A0A0P4XJM8 A0A1B0CL08 A0A158NU81 F4WR61 A0A182QD42 A0A084W1N9 A0A151WET4 A0A182NA85 T1E8R3 A0A1S3HUP4 A0A1S3HDP5 E9J2N0 A0A2K5ITY5 A0A2K5XB13 A0A096NL01 A0A2K5KTA2 A0A2K6QVL7 A0A2K6L3Y2 A0A151J377 A0A0D9R5A0 A0A182GCP5 A0A1S3WWM7 V4BZW1 A0A151IQF2 B0WS62 A0A2I2YE76 Q9D0K0 G1QJ66 A0A2R9CS18 A0A2I3T1E4 A0A2J8WN18 A0A024QZX0 Q9P0N9 G1T2E1 A0A2K6BL71 G7MQR4 G7P4F8 A0A1A8PLC4 A0A1A8QBX7 A0A195BAR2 A0A3Q0CJ09 A0A195FJL9 H9FVD2 A0A1A8KBR6 A0A1A8BI44 A0A1A8VIU1 D4AAY4
S4NXH8 A0A212FCI9 A0A194PLN9 A0A0L7LCJ7 A0A2P8YJ97 A0A2J7PT35 A0A067QXG3 D6X1P7 E0VFS3 A0A336N1M4 A0A2A3EEU0 N6TM20 J3JV08 A0A232FAS6 K7IXX5 A0A1B6C0Y6 E2AMK7 A0A026W5N2 E9GLF0 U5EYJ8 U4UIC9 A0A1B6K8B6 E2BJX5 A0A182M7E1 A0A1B0GMI1 A0A1Y1KZY2 A0A0T6BG58 A0A1W4XDZ7 A0A1Q3F2R1 A0A154P214 A0A2M4AS64 A0A182HT82 A0A0L7QQT5 A0A0P5K270 A0A182YKI3 A0A1C7CZV8 Q7QC09 A0A182JX80 A0A182J6Y1 A0A182V7N6 A0A182UC53 A0A182SXF7 A0A182PA60 A0A182VX56 W5JL26 A0A0P5NLA5 A0A182XI95 A0A0P4XJM8 A0A1B0CL08 A0A158NU81 F4WR61 A0A182QD42 A0A084W1N9 A0A151WET4 A0A182NA85 T1E8R3 A0A1S3HUP4 A0A1S3HDP5 E9J2N0 A0A2K5ITY5 A0A2K5XB13 A0A096NL01 A0A2K5KTA2 A0A2K6QVL7 A0A2K6L3Y2 A0A151J377 A0A0D9R5A0 A0A182GCP5 A0A1S3WWM7 V4BZW1 A0A151IQF2 B0WS62 A0A2I2YE76 Q9D0K0 G1QJ66 A0A2R9CS18 A0A2I3T1E4 A0A2J8WN18 A0A024QZX0 Q9P0N9 G1T2E1 A0A2K6BL71 G7MQR4 G7P4F8 A0A1A8PLC4 A0A1A8QBX7 A0A195BAR2 A0A3Q0CJ09 A0A195FJL9 H9FVD2 A0A1A8KBR6 A0A1A8BI44 A0A1A8VIU1 D4AAY4
Pubmed
19121390
28756777
26354079
23622113
22118469
26227816
+ More
29403074 24845553 18362917 19820115 20566863 23537049 22516182 28648823 20075255 20798317 24508170 30249741 21292972 28004739 25244985 20966253 12364791 14747013 17210077 20920257 23761445 21347285 21719571 24438588 21282665 25362486 26483478 23254933 22398555 16141072 15489334 21183079 22795129 22722832 16136131 11181995 19077034 14702039 11042152 14574404 17658474 23687350 24515783 21993624 22002653 25319552 15057822
29403074 24845553 18362917 19820115 20566863 23537049 22516182 28648823 20075255 20798317 24508170 30249741 21292972 28004739 25244985 20966253 12364791 14747013 17210077 20920257 23761445 21347285 21719571 24438588 21282665 25362486 26483478 23254933 22398555 16141072 15489334 21183079 22795129 22722832 16136131 11181995 19077034 14702039 11042152 14574404 17658474 23687350 24515783 21993624 22002653 25319552 15057822
EMBL
BABH01016525
KZ149908
PZC78219.1
NWSH01002624
PCG67823.1
ODYU01009043
+ More
SOQ53165.1 GDQN01001837 JAT89217.1 KQ461003 KPJ10026.1 GAIX01014195 JAA78365.1 AGBW02009182 OWR51454.1 KQ459601 KPI93649.1 JTDY01001702 KOB73120.1 PYGN01000555 PSN44323.1 NEVH01021921 PNF19492.1 KK853244 KDR09437.1 KQ971371 EFA10139.1 DS235124 EEB12229.1 UFQS01000739 UFQT01000739 UFQT01004060 SSX06490.1 SSX35469.1 KZ288271 PBC29792.1 APGK01058789 KB741292 ENN70300.1 BT127073 AEE62035.1 NNAY01000552 OXU27722.1 GEDC01030388 JAS06910.1 GL440824 EFN65327.1 KK107390 QOIP01000007 EZA51380.1 RLU20185.1 GL732551 EFX79502.1 GANO01001959 JAB57912.1 KB632334 ERL92772.1 GECU01000035 JAT07672.1 GL448708 EFN83984.1 AXCM01000651 AJVK01024552 GEZM01074853 JAV64457.1 LJIG01000658 KRT86282.1 GFDL01013228 JAV21817.1 KQ434803 KZC05975.1 GGFK01010325 MBW43646.1 APCN01000421 KQ414785 KOC60993.1 GDIQ01216409 JAK35316.1 AAAB01008859 EAA08097.4 ADMH02001252 ETN63459.1 GDIQ01162786 JAK88939.1 GDIP01242060 LRGB01000626 JAI81341.1 KZS17580.1 AJWK01016825 ADTU01026286 GL888284 EGI63288.1 AXCN02000218 ATLV01019399 KE525269 KFB44133.1 KQ983238 KYQ46360.1 GAMD01002543 JAA99047.1 GL767899 EFZ12926.1 AHZZ02024147 KQ980298 KYN16837.1 AQIB01159363 JXUM01054531 KQ561824 KXJ77422.1 KB201751 ESO94704.1 KQ976780 KYN08381.1 DS232065 EDS33685.1 CABD030043362 CABD030043363 CABD030043364 AK011356 AK156543 CH466546 BC125307 BC125309 BC145156 BC145157 ADFV01015625 ADFV01015626 AJFE02080831 AJFE02080832 AACZ04065916 GABC01008906 GABF01007189 GABD01001234 GABD01001233 GABE01005653 NBAG03000244 JAA02432.1 JAA14956.1 JAA31866.1 JAA39086.1 PNI62803.1 PNI62807.1 PNI62814.1 ABGA01346201 ABGA01346202 ABGA01346203 ABGA01346204 ABGA01346205 ABGA01346206 NDHI03003384 PNJ71168.1 PNJ71170.1 PNJ71171.1 CH471087 EAW55326.1 AB449888 AY826820 AK057228 AF151073 AY542308 AK223445 AL008729 AL589984 BC050465 BC007054 AAGW02052177 CM001256 EHH17960.1 AQIA01052735 AQIA01052736 CM001279 EHH52721.1 HAEI01002872 HAEH01012699 SBR82210.1 HAEF01008235 HAEG01012039 SBR91310.1 KQ976532 KYM81636.1 KQ981512 KYN40855.1 JU334838 JU475865 JV047269 AFE78591.1 AFH32669.1 AFI37340.1 HAED01022354 HAEE01009815 SBR29865.1 HADZ01003346 HAEA01014195 SBP67287.1 HADY01013685 HAEJ01019020 SBS59477.1 AABR07027309
SOQ53165.1 GDQN01001837 JAT89217.1 KQ461003 KPJ10026.1 GAIX01014195 JAA78365.1 AGBW02009182 OWR51454.1 KQ459601 KPI93649.1 JTDY01001702 KOB73120.1 PYGN01000555 PSN44323.1 NEVH01021921 PNF19492.1 KK853244 KDR09437.1 KQ971371 EFA10139.1 DS235124 EEB12229.1 UFQS01000739 UFQT01000739 UFQT01004060 SSX06490.1 SSX35469.1 KZ288271 PBC29792.1 APGK01058789 KB741292 ENN70300.1 BT127073 AEE62035.1 NNAY01000552 OXU27722.1 GEDC01030388 JAS06910.1 GL440824 EFN65327.1 KK107390 QOIP01000007 EZA51380.1 RLU20185.1 GL732551 EFX79502.1 GANO01001959 JAB57912.1 KB632334 ERL92772.1 GECU01000035 JAT07672.1 GL448708 EFN83984.1 AXCM01000651 AJVK01024552 GEZM01074853 JAV64457.1 LJIG01000658 KRT86282.1 GFDL01013228 JAV21817.1 KQ434803 KZC05975.1 GGFK01010325 MBW43646.1 APCN01000421 KQ414785 KOC60993.1 GDIQ01216409 JAK35316.1 AAAB01008859 EAA08097.4 ADMH02001252 ETN63459.1 GDIQ01162786 JAK88939.1 GDIP01242060 LRGB01000626 JAI81341.1 KZS17580.1 AJWK01016825 ADTU01026286 GL888284 EGI63288.1 AXCN02000218 ATLV01019399 KE525269 KFB44133.1 KQ983238 KYQ46360.1 GAMD01002543 JAA99047.1 GL767899 EFZ12926.1 AHZZ02024147 KQ980298 KYN16837.1 AQIB01159363 JXUM01054531 KQ561824 KXJ77422.1 KB201751 ESO94704.1 KQ976780 KYN08381.1 DS232065 EDS33685.1 CABD030043362 CABD030043363 CABD030043364 AK011356 AK156543 CH466546 BC125307 BC125309 BC145156 BC145157 ADFV01015625 ADFV01015626 AJFE02080831 AJFE02080832 AACZ04065916 GABC01008906 GABF01007189 GABD01001234 GABD01001233 GABE01005653 NBAG03000244 JAA02432.1 JAA14956.1 JAA31866.1 JAA39086.1 PNI62803.1 PNI62807.1 PNI62814.1 ABGA01346201 ABGA01346202 ABGA01346203 ABGA01346204 ABGA01346205 ABGA01346206 NDHI03003384 PNJ71168.1 PNJ71170.1 PNJ71171.1 CH471087 EAW55326.1 AB449888 AY826820 AK057228 AF151073 AY542308 AK223445 AL008729 AL589984 BC050465 BC007054 AAGW02052177 CM001256 EHH17960.1 AQIA01052735 AQIA01052736 CM001279 EHH52721.1 HAEI01002872 HAEH01012699 SBR82210.1 HAEF01008235 HAEG01012039 SBR91310.1 KQ976532 KYM81636.1 KQ981512 KYN40855.1 JU334838 JU475865 JV047269 AFE78591.1 AFH32669.1 AFI37340.1 HAED01022354 HAEE01009815 SBR29865.1 HADZ01003346 HAEA01014195 SBP67287.1 HADY01013685 HAEJ01019020 SBS59477.1 AABR07027309
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000245037 UP000235965 UP000027135 UP000007266 UP000009046 UP000242457 UP000019118 UP000215335 UP000002358 UP000000311 UP000053097 UP000279307 UP000000305 UP000030742 UP000008237 UP000075883 UP000092462 UP000192223 UP000076502 UP000075840 UP000053825 UP000076408 UP000075882 UP000007062 UP000075881 UP000075880 UP000075903 UP000075902 UP000075901 UP000075885 UP000075920 UP000000673 UP000076407 UP000076858 UP000092461 UP000005205 UP000007755 UP000075886 UP000030765 UP000075809 UP000075884 UP000085678 UP000233080 UP000233140 UP000028761 UP000233060 UP000233200 UP000233180 UP000078492 UP000029965 UP000069940 UP000249989 UP000079721 UP000030746 UP000078542 UP000002320 UP000001519 UP000000589 UP000001073 UP000240080 UP000002277 UP000001595 UP000005640 UP000001811 UP000233120 UP000009130 UP000233100 UP000078540 UP000189706 UP000078541 UP000002494
UP000245037 UP000235965 UP000027135 UP000007266 UP000009046 UP000242457 UP000019118 UP000215335 UP000002358 UP000000311 UP000053097 UP000279307 UP000000305 UP000030742 UP000008237 UP000075883 UP000092462 UP000192223 UP000076502 UP000075840 UP000053825 UP000076408 UP000075882 UP000007062 UP000075881 UP000075880 UP000075903 UP000075902 UP000075901 UP000075885 UP000075920 UP000000673 UP000076407 UP000076858 UP000092461 UP000005205 UP000007755 UP000075886 UP000030765 UP000075809 UP000075884 UP000085678 UP000233080 UP000233140 UP000028761 UP000233060 UP000233200 UP000233180 UP000078492 UP000029965 UP000069940 UP000249989 UP000079721 UP000030746 UP000078542 UP000002320 UP000001519 UP000000589 UP000001073 UP000240080 UP000002277 UP000001595 UP000005640 UP000001811 UP000233120 UP000009130 UP000233100 UP000078540 UP000189706 UP000078541 UP000002494
PRIDE
Interpro
CDD
ProteinModelPortal
H9JMN1
A0A2W1BT38
A0A2A4J8V2
A0A2H1WJB3
A0A1E1WQM1
A0A0N0PBG0
+ More
S4NXH8 A0A212FCI9 A0A194PLN9 A0A0L7LCJ7 A0A2P8YJ97 A0A2J7PT35 A0A067QXG3 D6X1P7 E0VFS3 A0A336N1M4 A0A2A3EEU0 N6TM20 J3JV08 A0A232FAS6 K7IXX5 A0A1B6C0Y6 E2AMK7 A0A026W5N2 E9GLF0 U5EYJ8 U4UIC9 A0A1B6K8B6 E2BJX5 A0A182M7E1 A0A1B0GMI1 A0A1Y1KZY2 A0A0T6BG58 A0A1W4XDZ7 A0A1Q3F2R1 A0A154P214 A0A2M4AS64 A0A182HT82 A0A0L7QQT5 A0A0P5K270 A0A182YKI3 A0A1C7CZV8 Q7QC09 A0A182JX80 A0A182J6Y1 A0A182V7N6 A0A182UC53 A0A182SXF7 A0A182PA60 A0A182VX56 W5JL26 A0A0P5NLA5 A0A182XI95 A0A0P4XJM8 A0A1B0CL08 A0A158NU81 F4WR61 A0A182QD42 A0A084W1N9 A0A151WET4 A0A182NA85 T1E8R3 A0A1S3HUP4 A0A1S3HDP5 E9J2N0 A0A2K5ITY5 A0A2K5XB13 A0A096NL01 A0A2K5KTA2 A0A2K6QVL7 A0A2K6L3Y2 A0A151J377 A0A0D9R5A0 A0A182GCP5 A0A1S3WWM7 V4BZW1 A0A151IQF2 B0WS62 A0A2I2YE76 Q9D0K0 G1QJ66 A0A2R9CS18 A0A2I3T1E4 A0A2J8WN18 A0A024QZX0 Q9P0N9 G1T2E1 A0A2K6BL71 G7MQR4 G7P4F8 A0A1A8PLC4 A0A1A8QBX7 A0A195BAR2 A0A3Q0CJ09 A0A195FJL9 H9FVD2 A0A1A8KBR6 A0A1A8BI44 A0A1A8VIU1 D4AAY4
S4NXH8 A0A212FCI9 A0A194PLN9 A0A0L7LCJ7 A0A2P8YJ97 A0A2J7PT35 A0A067QXG3 D6X1P7 E0VFS3 A0A336N1M4 A0A2A3EEU0 N6TM20 J3JV08 A0A232FAS6 K7IXX5 A0A1B6C0Y6 E2AMK7 A0A026W5N2 E9GLF0 U5EYJ8 U4UIC9 A0A1B6K8B6 E2BJX5 A0A182M7E1 A0A1B0GMI1 A0A1Y1KZY2 A0A0T6BG58 A0A1W4XDZ7 A0A1Q3F2R1 A0A154P214 A0A2M4AS64 A0A182HT82 A0A0L7QQT5 A0A0P5K270 A0A182YKI3 A0A1C7CZV8 Q7QC09 A0A182JX80 A0A182J6Y1 A0A182V7N6 A0A182UC53 A0A182SXF7 A0A182PA60 A0A182VX56 W5JL26 A0A0P5NLA5 A0A182XI95 A0A0P4XJM8 A0A1B0CL08 A0A158NU81 F4WR61 A0A182QD42 A0A084W1N9 A0A151WET4 A0A182NA85 T1E8R3 A0A1S3HUP4 A0A1S3HDP5 E9J2N0 A0A2K5ITY5 A0A2K5XB13 A0A096NL01 A0A2K5KTA2 A0A2K6QVL7 A0A2K6L3Y2 A0A151J377 A0A0D9R5A0 A0A182GCP5 A0A1S3WWM7 V4BZW1 A0A151IQF2 B0WS62 A0A2I2YE76 Q9D0K0 G1QJ66 A0A2R9CS18 A0A2I3T1E4 A0A2J8WN18 A0A024QZX0 Q9P0N9 G1T2E1 A0A2K6BL71 G7MQR4 G7P4F8 A0A1A8PLC4 A0A1A8QBX7 A0A195BAR2 A0A3Q0CJ09 A0A195FJL9 H9FVD2 A0A1A8KBR6 A0A1A8BI44 A0A1A8VIU1 D4AAY4
PDB
3QWL
E-value=2.60808e-55,
Score=544
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasmic vesicle
Localizes in the cytoplasmic vesicles of the endomembrane in association with TSC1-TSC2 complex. With evidence from 1 publications.
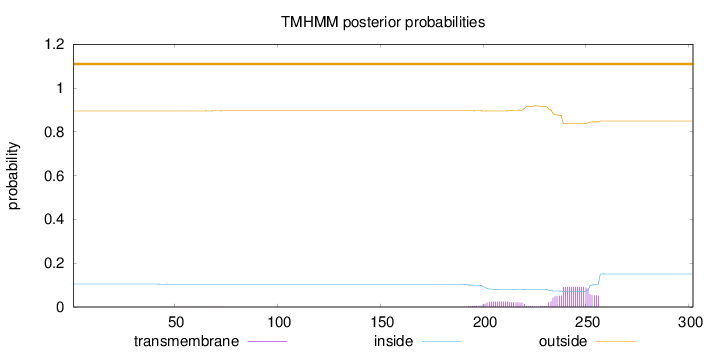
Length:
302
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.27673
Exp number, first 60 AAs:
0.01213
Total prob of N-in:
0.10468
outside
1 - 302
Population Genetic Test Statistics
Pi
225.660562
Theta
134.39946
Tajima's D
2.462869
CLR
0.312178
CSRT
0.939753012349383
Interpretation
Uncertain
