Gene
KWMTBOMO15476
Annotation
mariner_transposase_[Bombyx_mori]
Full name
Innexin
Location in the cell
Nuclear Reliability : 2.834
Sequence
CDS
ATGATTTCTGCATTTCGTGATGAAGCCCCATCCAAAACCACAATTTATCGCTGGTTTGCTGAGTTTCAACGTGGACGTGTCAAGCTCAGTGATGATCCCCGTCAAGGTCGTCCAAAAACTGCAGTCACCCAAGAAAACGTTGATGCTGTGCGTAAGCTGATTGAGGAAGATCGACATGTGACATACCGCGAAATTCAGGCAACTTTAGACATTGGCATGAGTCAAACACAAATAATCTTGCATGAACAATTAGGTGTAAAAAAGTTGTTTTCCCGATGGATACCGCATTCGCTCTGTGAAGAGCAAAAAGCGGCTCGCGTTACTTGGTGCGTCAGAACTCTCGAAAGATTCCACGCAGGATCCTCAAATGCTGTATACAACATTGTATCAGGTGACGAATCCTGGATATACGTGTACGAACCCGAAACAAAAAACCAGTCACGAGTTTGGGTGTTCGAAAATGAGTTAAAGCCAACAAAAATTGTTCGTTCACGGAGTGTTGCAAAAAAAATGGTGGCCACGTTTGTCTCCAAAACCGGCCATGTTACGACTATTCCTCTTGAGGGACAAAGAACGGTTAATGCAGAATGGTATGCTAGCATTTGTTTGCCACAGGTCGTTTCTGAACTCCGTAAAGAGAACTGCAACCGCCGCATCATCCTCCATCACGACAATGCGAGTTCTCACACCGCGCACAGAACAAAAGAGTTTTTAGAGCAAGAAAACATAGAATTATTAGACCATCCGCCGTACAGCCCCGACCTAAGCCCTAATGATTTCTATACTTTCCCTAAAATAAAGAATAAATTGCGTGGACAGAGATTTTCATCACCTGAAGAAGCTGTGGACGCCTACAAAACGGCCATTTTGGAGACCCCAACTTCCGAATGGAATGGTTGCTTCAATGATTGGTTCCATCGTATGGAAAAATGA
Protein
MISAFRDEAPSKTTIYRWFAEFQRGRVKLSDDPRQGRPKTAVTQENVDAVRKLIEEDRHVTYREIQATLDIGMSQTQIILHEQLGVKKLFSRWIPHSLCEEQKAARVTWCVRTLERFHAGSSNAVYNIVSGDESWIYVYEPETKNQSRVWVFENELKPTKIVRSRSVAKKMVATFVSKTGHVTTIPLEGQRTVNAEWYASICLPQVVSELRKENCNRRIILHHDNASSHTAHRTKEFLEQENIELLDHPPYSPDLSPNDFYTFPKIKNKLRGQRFSSPEEAVDAYKTAILETPTSEWNGCFNDWFHRMEK
Summary
Description
Structural component of the gap junctions.
Similarity
Belongs to the pannexin family.
Belongs to the histone H2A family.
Belongs to the histone H2A family.
Uniprot
Q1HPJ3
A0A023F6H6
A0A069DT60
A0A0V0G5J0
A0A1L5BY10
A0A034VN11
+ More
A0A267ESW3 A0A1B6JMK6 Q0QXC1 A0A267EV83 B3CK19 Q13539 B3CK17 B3CK18 A6GV71 B7ZDI8 B7ZDI6 B7ZDI7 A0A1L5BY07 A6GV70 A6GV69 B3CK26 A6GV72 B3CK24 A0A2H9T3D9 B3CK30 V5GGS5 A0A0J7MQ55 A0A267GHL5 B3CK23 A0A2J7R1D0 A0A1L5BY05 A0A1L5BY09 A0A2J7PES0 A0A2J7RSZ3 B3CK29 B3CK27 R4G5I0 A0A0J7NFU3 A0A2J7Q9R7 A0A0J7JXI0 A0A224XT40 A0A1L5BY08 A0A023F6P2 A0A023F6Y7 A0A1B6HD45 A0A3S1BW17 A0A2J7Q027 A0A267GB40 A0A1L5BXZ8 A0A267FT86 A0A2J7RMJ0 A0A069DS91 B3CK20 A0A224XRB4 A0A3S1B9R5 A0A085MI35 A0A085MXW7 A0A085MM23 A0A0K2UGY4 R4FP54 R4G4T8 A0A085MGE6 A0A085NH36 D2WLV0 A0A085LW38 Q04514 A0A069DT95 A0A069DTF4 A0A0J7K928 A0A1L8EJC7 V9H0F7 B3CK21 A0A1I8HRU7 A0A2J7Q4H1 A0A2J7PHE2 A0A1L5BY11 A0A085N4E0 A0A085N9Z9 A0A3Q0RHJ7 A0A1L8EGN5 A0A1L8E705 V9H0A8 A0A0J7MT17 R4FPS5 A0A3S0Z1J4 B3CK22 A0A267E0K0 A0A0J7KB36 A0A3S1A274
A0A267ESW3 A0A1B6JMK6 Q0QXC1 A0A267EV83 B3CK19 Q13539 B3CK17 B3CK18 A6GV71 B7ZDI8 B7ZDI6 B7ZDI7 A0A1L5BY07 A6GV70 A6GV69 B3CK26 A6GV72 B3CK24 A0A2H9T3D9 B3CK30 V5GGS5 A0A0J7MQ55 A0A267GHL5 B3CK23 A0A2J7R1D0 A0A1L5BY05 A0A1L5BY09 A0A2J7PES0 A0A2J7RSZ3 B3CK29 B3CK27 R4G5I0 A0A0J7NFU3 A0A2J7Q9R7 A0A0J7JXI0 A0A224XT40 A0A1L5BY08 A0A023F6P2 A0A023F6Y7 A0A1B6HD45 A0A3S1BW17 A0A2J7Q027 A0A267GB40 A0A1L5BXZ8 A0A267FT86 A0A2J7RMJ0 A0A069DS91 B3CK20 A0A224XRB4 A0A3S1B9R5 A0A085MI35 A0A085MXW7 A0A085MM23 A0A0K2UGY4 R4FP54 R4G4T8 A0A085MGE6 A0A085NH36 D2WLV0 A0A085LW38 Q04514 A0A069DT95 A0A069DTF4 A0A0J7K928 A0A1L8EJC7 V9H0F7 B3CK21 A0A1I8HRU7 A0A2J7Q4H1 A0A2J7PHE2 A0A1L5BY11 A0A085N4E0 A0A085N9Z9 A0A3Q0RHJ7 A0A1L8EGN5 A0A1L8E705 V9H0A8 A0A0J7MT17 R4FPS5 A0A3S0Z1J4 B3CK22 A0A267E0K0 A0A0J7KB36 A0A3S1A274
Pubmed
EMBL
DQ443409
ABF51498.1
GBBI01001865
JAC16847.1
GBGD01001983
JAC86906.1
+ More
GECL01002811 JAP03313.1 KX930998 APL98291.1 GAKP01015435 JAC43517.1 NIVC01001798 PAA63957.1 GECU01007301 JAT00406.1 DQ174779 ABA55502.1 NIVC01001731 PAA64652.1 AM906136 CAP20050.1 U49974 AAC52011.1 AM906133 CAP20047.1 AM906135 CAP20049.1 AM231072 CAJ76986.1 AM231082 CAJ76996.1 AM231079 CAJ76993.1 AM231080 CAJ76994.1 KX931001 APL98294.1 AM231070 CAJ76984.1 AM231069 CAJ76983.1 AM906145 CAP20059.1 AM231074 CAJ76988.1 AM906142 AM906144 CAP20056.1 NSIT01000395 PJE77748.1 AM906154 CAP20068.1 GALX01005282 JAB63184.1 LBMM01023556 KMQ82700.1 NIVC01000364 PAA84702.1 AM906141 CAP20055.1 NEVH01008208 PNF34637.1 KX930994 APL98287.1 KX930999 APL98292.1 NEVH01027114 NEVH01027075 NEVH01026089 NEVH01026087 NEVH01024426 NEVH01021956 NEVH01021925 NEVH01015301 NEVH01013964 NEVH01013194 NEVH01011885 NEVH01009393 NEVH01007824 NEVH01007823 NEVH01006721 NEVH01006578 NEVH01005888 NEVH01005885 NEVH01005277 NEVH01002149 NEVH01001358 PNF13652.1 PNF13785.1 PNF14826.1 PNF14870.1 PNF17130.1 PNF18970.1 PNF19346.1 PNF27270.1 PNF28165.1 PNF28197.1 PNF30137.1 PNF31284.1 PNF33174.1 PNF35196.1 PNF35254.1 PNF37314.1 PNF37715.1 PNF38363.1 PNF38519.1 PNF39139.1 PNF42450.1 PNF42722.1 NEVH01000249 PNF43951.1 AM906153 CAP20067.1 AM906151 AM906152 CAP20065.1 GAHY01000550 JAA76960.1 LBMM01005538 KMQ91440.1 NEVH01020956 NEVH01016344 NEVH01006723 NEVH01003746 PNF20350.1 PNF25331.1 PNF37025.1 PNF40184.1 LBMM01023216 KMQ82764.1 GFTR01005295 JAW11131.1 KX931000 APL98293.1 GBBI01001812 JAC16900.1 GBBI01001710 JAC17002.1 GECU01035151 JAS72555.1 RQTK01000675 RUS76288.1 NEVH01020327 PNF21945.1 NIVC01001708 NIVC01001487 NIVC01000426 NIVC01000107 PAA64861.1 PAA67372.1 PAA83241.1 PAA90594.1 KX930995 APL98288.1 NIVC01000831 PAA76287.1 NEVH01002553 PNF42057.1 GBGD01002089 JAC86800.1 AM906137 CAP20051.1 GFTR01005334 JAW11092.1 RQTK01000506 RUS78489.1 KL363191 KFD56881.1 KL367603 KFD62063.1 KL363185 KFD58269.1 HACA01020102 CDW37463.1 GAHY01001054 JAA76456.1 GAHY01001080 JAA76430.1 KL363194 KFD56292.1 KL367501 KFD68782.1 GQ398105 ADB28039.1 KL363276 KFD49184.1 L06041 U11652 AAA28265.1 AAC46947.1 GBGD01001923 JAC86966.1 GBGD01001947 JAC86942.1 LBMM01011272 KMQ86923.1 GFDG01000031 JAV18768.1 U11650 AAC46945.1 AM906139 CAP20053.1 NEVH01018383 PNF23469.1 NEVH01025141 PNF15743.1 KX931003 APL98296.1 KL367557 KFD64336.1 KL367526 KFD66295.1 GFDG01000909 JAV17890.1 GFDG01004365 JAV14434.1 U11654 AAC46948.1 LBMM01019009 KMQ83645.1 GAHY01000734 JAA76776.1 RQTK01002074 RUS68713.1 AM906140 CAP20054.1 NIVC01002911 PAA54417.1 LBMM01010241 KMQ87583.1 RQTK01000075 RUS88698.1
GECL01002811 JAP03313.1 KX930998 APL98291.1 GAKP01015435 JAC43517.1 NIVC01001798 PAA63957.1 GECU01007301 JAT00406.1 DQ174779 ABA55502.1 NIVC01001731 PAA64652.1 AM906136 CAP20050.1 U49974 AAC52011.1 AM906133 CAP20047.1 AM906135 CAP20049.1 AM231072 CAJ76986.1 AM231082 CAJ76996.1 AM231079 CAJ76993.1 AM231080 CAJ76994.1 KX931001 APL98294.1 AM231070 CAJ76984.1 AM231069 CAJ76983.1 AM906145 CAP20059.1 AM231074 CAJ76988.1 AM906142 AM906144 CAP20056.1 NSIT01000395 PJE77748.1 AM906154 CAP20068.1 GALX01005282 JAB63184.1 LBMM01023556 KMQ82700.1 NIVC01000364 PAA84702.1 AM906141 CAP20055.1 NEVH01008208 PNF34637.1 KX930994 APL98287.1 KX930999 APL98292.1 NEVH01027114 NEVH01027075 NEVH01026089 NEVH01026087 NEVH01024426 NEVH01021956 NEVH01021925 NEVH01015301 NEVH01013964 NEVH01013194 NEVH01011885 NEVH01009393 NEVH01007824 NEVH01007823 NEVH01006721 NEVH01006578 NEVH01005888 NEVH01005885 NEVH01005277 NEVH01002149 NEVH01001358 PNF13652.1 PNF13785.1 PNF14826.1 PNF14870.1 PNF17130.1 PNF18970.1 PNF19346.1 PNF27270.1 PNF28165.1 PNF28197.1 PNF30137.1 PNF31284.1 PNF33174.1 PNF35196.1 PNF35254.1 PNF37314.1 PNF37715.1 PNF38363.1 PNF38519.1 PNF39139.1 PNF42450.1 PNF42722.1 NEVH01000249 PNF43951.1 AM906153 CAP20067.1 AM906151 AM906152 CAP20065.1 GAHY01000550 JAA76960.1 LBMM01005538 KMQ91440.1 NEVH01020956 NEVH01016344 NEVH01006723 NEVH01003746 PNF20350.1 PNF25331.1 PNF37025.1 PNF40184.1 LBMM01023216 KMQ82764.1 GFTR01005295 JAW11131.1 KX931000 APL98293.1 GBBI01001812 JAC16900.1 GBBI01001710 JAC17002.1 GECU01035151 JAS72555.1 RQTK01000675 RUS76288.1 NEVH01020327 PNF21945.1 NIVC01001708 NIVC01001487 NIVC01000426 NIVC01000107 PAA64861.1 PAA67372.1 PAA83241.1 PAA90594.1 KX930995 APL98288.1 NIVC01000831 PAA76287.1 NEVH01002553 PNF42057.1 GBGD01002089 JAC86800.1 AM906137 CAP20051.1 GFTR01005334 JAW11092.1 RQTK01000506 RUS78489.1 KL363191 KFD56881.1 KL367603 KFD62063.1 KL363185 KFD58269.1 HACA01020102 CDW37463.1 GAHY01001054 JAA76456.1 GAHY01001080 JAA76430.1 KL363194 KFD56292.1 KL367501 KFD68782.1 GQ398105 ADB28039.1 KL363276 KFD49184.1 L06041 U11652 AAA28265.1 AAC46947.1 GBGD01001923 JAC86966.1 GBGD01001947 JAC86942.1 LBMM01011272 KMQ86923.1 GFDG01000031 JAV18768.1 U11650 AAC46945.1 AM906139 CAP20053.1 NEVH01018383 PNF23469.1 NEVH01025141 PNF15743.1 KX931003 APL98296.1 KL367557 KFD64336.1 KL367526 KFD66295.1 GFDG01000909 JAV17890.1 GFDG01004365 JAV14434.1 U11654 AAC46948.1 LBMM01019009 KMQ83645.1 GAHY01000734 JAA76776.1 RQTK01002074 RUS68713.1 AM906140 CAP20054.1 NIVC01002911 PAA54417.1 LBMM01010241 KMQ87583.1 RQTK01000075 RUS88698.1
Proteomes
Pfam
Interpro
SUPFAM
SSF47113
SSF47113
Gene 3D
ProteinModelPortal
Q1HPJ3
A0A023F6H6
A0A069DT60
A0A0V0G5J0
A0A1L5BY10
A0A034VN11
+ More
A0A267ESW3 A0A1B6JMK6 Q0QXC1 A0A267EV83 B3CK19 Q13539 B3CK17 B3CK18 A6GV71 B7ZDI8 B7ZDI6 B7ZDI7 A0A1L5BY07 A6GV70 A6GV69 B3CK26 A6GV72 B3CK24 A0A2H9T3D9 B3CK30 V5GGS5 A0A0J7MQ55 A0A267GHL5 B3CK23 A0A2J7R1D0 A0A1L5BY05 A0A1L5BY09 A0A2J7PES0 A0A2J7RSZ3 B3CK29 B3CK27 R4G5I0 A0A0J7NFU3 A0A2J7Q9R7 A0A0J7JXI0 A0A224XT40 A0A1L5BY08 A0A023F6P2 A0A023F6Y7 A0A1B6HD45 A0A3S1BW17 A0A2J7Q027 A0A267GB40 A0A1L5BXZ8 A0A267FT86 A0A2J7RMJ0 A0A069DS91 B3CK20 A0A224XRB4 A0A3S1B9R5 A0A085MI35 A0A085MXW7 A0A085MM23 A0A0K2UGY4 R4FP54 R4G4T8 A0A085MGE6 A0A085NH36 D2WLV0 A0A085LW38 Q04514 A0A069DT95 A0A069DTF4 A0A0J7K928 A0A1L8EJC7 V9H0F7 B3CK21 A0A1I8HRU7 A0A2J7Q4H1 A0A2J7PHE2 A0A1L5BY11 A0A085N4E0 A0A085N9Z9 A0A3Q0RHJ7 A0A1L8EGN5 A0A1L8E705 V9H0A8 A0A0J7MT17 R4FPS5 A0A3S0Z1J4 B3CK22 A0A267E0K0 A0A0J7KB36 A0A3S1A274
A0A267ESW3 A0A1B6JMK6 Q0QXC1 A0A267EV83 B3CK19 Q13539 B3CK17 B3CK18 A6GV71 B7ZDI8 B7ZDI6 B7ZDI7 A0A1L5BY07 A6GV70 A6GV69 B3CK26 A6GV72 B3CK24 A0A2H9T3D9 B3CK30 V5GGS5 A0A0J7MQ55 A0A267GHL5 B3CK23 A0A2J7R1D0 A0A1L5BY05 A0A1L5BY09 A0A2J7PES0 A0A2J7RSZ3 B3CK29 B3CK27 R4G5I0 A0A0J7NFU3 A0A2J7Q9R7 A0A0J7JXI0 A0A224XT40 A0A1L5BY08 A0A023F6P2 A0A023F6Y7 A0A1B6HD45 A0A3S1BW17 A0A2J7Q027 A0A267GB40 A0A1L5BXZ8 A0A267FT86 A0A2J7RMJ0 A0A069DS91 B3CK20 A0A224XRB4 A0A3S1B9R5 A0A085MI35 A0A085MXW7 A0A085MM23 A0A0K2UGY4 R4FP54 R4G4T8 A0A085MGE6 A0A085NH36 D2WLV0 A0A085LW38 Q04514 A0A069DT95 A0A069DTF4 A0A0J7K928 A0A1L8EJC7 V9H0F7 B3CK21 A0A1I8HRU7 A0A2J7Q4H1 A0A2J7PHE2 A0A1L5BY11 A0A085N4E0 A0A085N9Z9 A0A3Q0RHJ7 A0A1L8EGN5 A0A1L8E705 V9H0A8 A0A0J7MT17 R4FPS5 A0A3S0Z1J4 B3CK22 A0A267E0K0 A0A0J7KB36 A0A3S1A274
PDB
5HOO
E-value=7.38941e-27,
Score=298
Ontologies
PANTHER
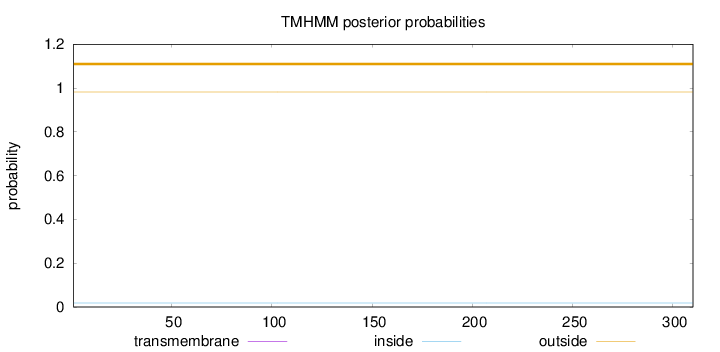
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cell junction
Gap junction
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000500000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01860
outside
1 - 310
Population Genetic Test Statistics
Pi
1.146179
Theta
1.551129
Tajima's D
-0.904473
CLR
111.503106
CSRT
0.160641967901605
Interpretation
Uncertain
