Gene
KWMTBOMO15468
Pre Gene Modal
BGIBMGA010774
Annotation
PREDICTED:_protein_Skeletor?_isoforms_B/C_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.566
Sequence
CDS
ATGTTATGTATTGGTTTCATCTTGTTCATAGGACTTGCTAAAGAATGTTATGCGCAGGATGATAATGGTCCCTACCTAGGACGATCTATTGGCGAGTTTAATACTTACCACCACCAGGTATCAGGTGATGTGTATGCTGTAGATGACTGGACCCTATTACTGGTGAATTTTAACTATGATGGTACTGGAGTTGACACATTCTTCTGGGCTGGAGATTCTGGGAGGCCAGGCCCACAGGGTTTTATAATCCCTGATCAATTTGGCAAAACCAATGTCCTGGAGCGATACTACAATGAGGAAGTACGACTCACACTTCCAGAAGGCAAGCGGATATCCCGAATCAAGTGGTTTGCCGTTTATGATATAGACTCACAGAATGCATTTGGGGATGTGTATATACCTGAAGATTTCTCTGCTCCAGCACCTCAGTCACTAGCAGCATTCTCCTCTGCTGCTGTGACAAGTAAACCTCTGAGAGTACTTGACGCTTCTACCATATTAATACCAGAATTCAGATACGACGGCCGTGGAGAAGAAGCCTACTTTTGGACGGGAGTCGGGCCACAGCCTTCATCACGAGGCTTCAAAATACCTGATGAATATGGATACCTGGAGCCAATCCGCGCGTACAAGGGCGAAGACGTGCGGCTAGAGCTCCCGGGCGGCAAGACCATATTCGACGTGAACTGGATCTCCGTGTACGACGCGTCCGCGCGCGCCAGCCTCGGCGCCGTGCTCGTGCCCGACGGACTCAACGTGCCGCCCGCCGCCCTCGTCACGCACCCGCACGAGAGCAAGTTGAGTCAGTGCAAGCAGCTGCACCGCGACCTCCAGGTCTCGTGGAACGTCCACGGCAGGGAGATCACCATCGAACTCGCGGGACAGATAGAGGAGGACGAGTACATGTCGTTCGGCATCTCCGGCGCGGCGGAGCGCTCCGCCATGCTCGGCGCCGACGTGGCGGTCGCGCGCTACTCCCTCGCAGAGAGGCGCGGCCTCGTCACCGACTACAACATCTCGGACGTGGCGCCGTGCGTGCGCGTGCTGGGCGTGTGGCGCGGCGTGTGTCGCGACACGGCGGTGGGCGGCCTGGACTCCCCGCAGCTGCTGTACGCGTCCCGGGAGGACTCTCTCACGCGCATCACCTACCGCACCAACCAGCAGCCCGACGAGGCTCTGGACCGAGAGTGGAAAAAAGACGGAGACCTTTATGTAGTGTGGGCTGTGGGGAAGCTGGACTCCCTCATGGAGCCCGCCTTCCATCGCCTATATCCGCGCCACGATGTCATCATACGCCTCAACAACACTCTGCCGGATAATGACTGTTTCAAGTTCACCATCGGCAGCAGGGCGCGGCCGAGTCCGTGGGAGGTGGCGCAACTGTTCGACCCCACCCTGCGCGTGTTCCAGTTCCGCGTGGGGCCGGCGGGGGGCGCGCGGGGGGTGGCGGGGCGCGCGGAGGGGCACGCGCCCCCCCTCGCCTGGTACGTCAACGGGCAGCTCGCGCCCGACATCCAGCTGCGCCGCGGACTCACCTACACCTTCTACGTGTACGGGGGCAACGAGCCCCACTCTGCCACTGAGTACCACCCGCTGGTGGTGACGGCGGAGCAGGGCGGCGGCCTGGAGCGCCTCTCGGACTCCGCCCTCCGACAAGTGCGCGTGCTCGCCGGCCTGCACTATTCGCGCCGCGGCCTGCTGCAGCCCACGGCCGCGGGCGGGCTGTGCCTGGGCCGCGCGGCGGAGGAGGCGGACCGGCGGCGGGACGCGGACTACGTGACGTTCCGCGCGTACAACAGGTCGCTGCGGTGGGAGTGCGCGTCGGCGAGCCCGGCCGCGCTCACGGTGTCCCCGAACTCGTCGTGGCCGGACATCGTGTACTACAACTCCTTCACGCACGCAGGCATGGGCGGCCGCATCTACATCGTGGACCGACACAACCGGAACATCGCCCGCAAGAGCGCCGCCGCGACGCTCGCCCCCCTCACCCCCCTCACCGTGCTCACGGCGCTCACCGTGCTCACCGTGCTCACCGCGCTCACGGCCCGGTGA
Protein
MLCIGFILFIGLAKECYAQDDNGPYLGRSIGEFNTYHHQVSGDVYAVDDWTLLLVNFNYDGTGVDTFFWAGDSGRPGPQGFIIPDQFGKTNVLERYYNEEVRLTLPEGKRISRIKWFAVYDIDSQNAFGDVYIPEDFSAPAPQSLAAFSSAAVTSKPLRVLDASTILIPEFRYDGRGEEAYFWTGVGPQPSSRGFKIPDEYGYLEPIRAYKGEDVRLELPGGKTIFDVNWISVYDASARASLGAVLVPDGLNVPPAALVTHPHESKLSQCKQLHRDLQVSWNVHGREITIELAGQIEEDEYMSFGISGAAERSAMLGADVAVARYSLAERRGLVTDYNISDVAPCVRVLGVWRGVCRDTAVGGLDSPQLLYASREDSLTRITYRTNQQPDEALDREWKKDGDLYVVWAVGKLDSLMEPAFHRLYPRHDVIIRLNNTLPDNDCFKFTIGSRARPSPWEVAQLFDPTLRVFQFRVGPAGGARGVAGRAEGHAPPLAWYVNGQLAPDIQLRRGLTYTFYVYGGNEPHSATEYHPLVVTAEQGGGLERLSDSALRQVRVLAGLHYSRRGLLQPTAAGGLCLGRAAEEADRRRDADYVTFRAYNRSLRWECASASPAALTVSPNSSWPDIVYYNSFTHAGMGGRIYIVDRHNRNIARKSAAATLAPLTPLTVLTALTVLTVLTALTAR
Summary
Uniprot
A0A212EJ64
A0A2H1WTX1
E0VVB8
A0A194PJN3
A0A2W1BBJ5
A0A1S4G0S6
+ More
Q16HF5 A0A1B0CTV7 A0A2M4AAL0 A0A182G7Z6 A0A1B0DQ19 W5JAV9 A0A182PP74 A0A2M4BFU1 A0A0M4EVW7 A0A2M4BFL6 A0A182FAN7 A0A182QZ00 A0A182MQA6 A0A2M3Z490 A0A182W3G3 A0A182SPI1 A0A0L7QL47 A0A182NGF9 A0A2M3ZE23 A0A182JDJ3 A0A182Y8S1 A0A084WP56 A0A182RE39 A0A182K2Q5 A0A182XII5 A0A182HYP4 A0A182VMM3 Q7QA02 A0A1Y1L388 B0X1J7 A0A182KZJ3 D6X2I0 A0A2J7PYU2 A0A154P9T0 A0A0A1XEJ8 A0A0K8VNI7 A0A034VLB8 A0A1I8P161 A0A0R1E237 B4LXU7 A0A182TDM7 A0A1I8N0G7 B4K5C5 F4WUV5 W8B8E7 T1PQ56 B4PKL6 A0A0C9RT20 A0A0L0CJP2 B4G6D2 A0A1B0FN49 A0A1B6LC76 B4HJ91 Q9VH23 B4QVB1 A0A3B0KNI3 B3P4K8 B4JTX3 B3LWS7 B4NA47 A0A195CXL5 A0A1W4W076 A0A195ATX6 A0A1W4VZC8 E2AKF2 A0A1J1HK80 A0A151JZ33 A0A151WW69 A0A158P0U0 A0A336LTC4 A0A0M8ZW79 A0A069DVL9 A0A146M150 A0A0A9XRG9 A0A1B0BWG6 A0A026WK81 A0A1B0A7W4 T1HWC2 A0A1A9V8S2 A0A2A3EUM4 A0A2R7WUS0 A0A088AF18 A0A1B6DSH2 A0A1A9WNN2 K7IVC6 I5ANC2 E2BE34 A0A067R723 A0A232EIM5 A0A2J7PYQ9 A0A0P5I2A2 A0A0P5HBI7 A0A0P5FQZ2
Q16HF5 A0A1B0CTV7 A0A2M4AAL0 A0A182G7Z6 A0A1B0DQ19 W5JAV9 A0A182PP74 A0A2M4BFU1 A0A0M4EVW7 A0A2M4BFL6 A0A182FAN7 A0A182QZ00 A0A182MQA6 A0A2M3Z490 A0A182W3G3 A0A182SPI1 A0A0L7QL47 A0A182NGF9 A0A2M3ZE23 A0A182JDJ3 A0A182Y8S1 A0A084WP56 A0A182RE39 A0A182K2Q5 A0A182XII5 A0A182HYP4 A0A182VMM3 Q7QA02 A0A1Y1L388 B0X1J7 A0A182KZJ3 D6X2I0 A0A2J7PYU2 A0A154P9T0 A0A0A1XEJ8 A0A0K8VNI7 A0A034VLB8 A0A1I8P161 A0A0R1E237 B4LXU7 A0A182TDM7 A0A1I8N0G7 B4K5C5 F4WUV5 W8B8E7 T1PQ56 B4PKL6 A0A0C9RT20 A0A0L0CJP2 B4G6D2 A0A1B0FN49 A0A1B6LC76 B4HJ91 Q9VH23 B4QVB1 A0A3B0KNI3 B3P4K8 B4JTX3 B3LWS7 B4NA47 A0A195CXL5 A0A1W4W076 A0A195ATX6 A0A1W4VZC8 E2AKF2 A0A1J1HK80 A0A151JZ33 A0A151WW69 A0A158P0U0 A0A336LTC4 A0A0M8ZW79 A0A069DVL9 A0A146M150 A0A0A9XRG9 A0A1B0BWG6 A0A026WK81 A0A1B0A7W4 T1HWC2 A0A1A9V8S2 A0A2A3EUM4 A0A2R7WUS0 A0A088AF18 A0A1B6DSH2 A0A1A9WNN2 K7IVC6 I5ANC2 E2BE34 A0A067R723 A0A232EIM5 A0A2J7PYQ9 A0A0P5I2A2 A0A0P5HBI7 A0A0P5FQZ2
Pubmed
22118469
20566863
26354079
28756777
17510324
26483478
+ More
20920257 23761445 25244985 24438588 12364791 14747013 17210077 28004739 20966253 18362917 19820115 21930896 25830018 25348373 17994087 17550304 25315136 21719571 24495485 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 21347285 26334808 26823975 25401762 24508170 20075255 15632085 24845553 28648823
20920257 23761445 25244985 24438588 12364791 14747013 17210077 28004739 20966253 18362917 19820115 21930896 25830018 25348373 17994087 17550304 25315136 21719571 24495485 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 21347285 26334808 26823975 25401762 24508170 20075255 15632085 24845553 28648823
EMBL
AGBW02014522
OWR41526.1
ODYU01011036
SOQ56519.1
DS235806
EEB17324.1
+ More
KQ459601 KPI93636.1 KZ150485 PZC70707.1 CH478177 EAT33677.1 AJWK01028036 GGFK01004502 MBW37823.1 JXUM01151241 KQ571173 KXJ68231.1 AJVK01018738 ADMH02002016 ETN59980.1 GGFJ01002690 MBW51831.1 CP012526 ALC47567.1 GGFJ01002691 MBW51832.1 AXCN02001607 AXCM01001003 GGFM01002586 MBW23337.1 KQ414940 KOC59246.1 GGFM01006053 MBW26804.1 ATLV01024911 ATLV01024912 KE525364 KFB52000.1 APCN01005395 AAAB01008898 EAA09270.5 GEZM01066229 JAV68044.1 DS232261 EDS38646.1 JN314843 KQ971372 AEM60136.1 EFA09441.1 NEVH01020342 PNF21472.1 KQ434851 KZC08593.1 GBXI01004900 JAD09392.1 GDHF01025192 GDHF01016038 GDHF01011901 JAI27122.1 JAI36276.1 JAI40413.1 GAKP01014846 JAC44106.1 CM000160 KRK03325.1 CH940650 EDW67906.1 CH933806 EDW16151.2 GL888378 EGI62019.1 GAMC01009061 GAMC01009060 GAMC01009059 JAB97495.1 KA650253 AFP64882.1 EDW96775.1 GBYB01011787 GBYB01011788 GBYB01011789 GBYB01011790 JAG81554.1 JAG81555.1 JAG81556.1 JAG81557.1 JRES01000310 KNC32422.1 CH479179 EDW23890.1 CCAG010020972 GEBQ01018689 JAT21288.1 CH480815 EDW42763.1 AE014297 AY071192 AAF54497.1 AAL48814.1 CM000364 EDX13517.1 OUUW01000013 SPP88159.1 CH954181 EDV49661.1 CH916374 EDV91552.1 CH902617 EDV42715.1 CH964232 EDW80690.2 KQ977141 KYN05386.1 KQ976738 KYM75683.1 GL440262 EFN66094.1 CVRI01000002 CRK86870.1 KQ981414 KYN41868.1 KQ982691 KYQ52078.1 ADTU01005791 UFQT01000111 SSX20191.1 KQ435821 KOX72375.1 GBGD01000741 JAC88148.1 GDHC01005088 JAQ13541.1 GBHO01022156 GBRD01008035 JAG21448.1 JAG57786.1 JXJN01021770 KK107199 EZA55509.1 ACPB03016234 KZ288185 PBC34916.1 KK855619 PTY23302.1 GEDC01008695 JAS28603.1 AAZX01001195 CM000070 EIM52457.2 GL447755 EFN86021.1 KK852660 KDR19130.1 NNAY01004201 OXU18206.1 PNF21473.1 GDIQ01242201 GDIQ01210192 JAK09524.1 GDIQ01229038 JAK22687.1 GDIP01144919 GDIP01058394 LRGB01001361 JAJ78483.1 KZS12519.1
KQ459601 KPI93636.1 KZ150485 PZC70707.1 CH478177 EAT33677.1 AJWK01028036 GGFK01004502 MBW37823.1 JXUM01151241 KQ571173 KXJ68231.1 AJVK01018738 ADMH02002016 ETN59980.1 GGFJ01002690 MBW51831.1 CP012526 ALC47567.1 GGFJ01002691 MBW51832.1 AXCN02001607 AXCM01001003 GGFM01002586 MBW23337.1 KQ414940 KOC59246.1 GGFM01006053 MBW26804.1 ATLV01024911 ATLV01024912 KE525364 KFB52000.1 APCN01005395 AAAB01008898 EAA09270.5 GEZM01066229 JAV68044.1 DS232261 EDS38646.1 JN314843 KQ971372 AEM60136.1 EFA09441.1 NEVH01020342 PNF21472.1 KQ434851 KZC08593.1 GBXI01004900 JAD09392.1 GDHF01025192 GDHF01016038 GDHF01011901 JAI27122.1 JAI36276.1 JAI40413.1 GAKP01014846 JAC44106.1 CM000160 KRK03325.1 CH940650 EDW67906.1 CH933806 EDW16151.2 GL888378 EGI62019.1 GAMC01009061 GAMC01009060 GAMC01009059 JAB97495.1 KA650253 AFP64882.1 EDW96775.1 GBYB01011787 GBYB01011788 GBYB01011789 GBYB01011790 JAG81554.1 JAG81555.1 JAG81556.1 JAG81557.1 JRES01000310 KNC32422.1 CH479179 EDW23890.1 CCAG010020972 GEBQ01018689 JAT21288.1 CH480815 EDW42763.1 AE014297 AY071192 AAF54497.1 AAL48814.1 CM000364 EDX13517.1 OUUW01000013 SPP88159.1 CH954181 EDV49661.1 CH916374 EDV91552.1 CH902617 EDV42715.1 CH964232 EDW80690.2 KQ977141 KYN05386.1 KQ976738 KYM75683.1 GL440262 EFN66094.1 CVRI01000002 CRK86870.1 KQ981414 KYN41868.1 KQ982691 KYQ52078.1 ADTU01005791 UFQT01000111 SSX20191.1 KQ435821 KOX72375.1 GBGD01000741 JAC88148.1 GDHC01005088 JAQ13541.1 GBHO01022156 GBRD01008035 JAG21448.1 JAG57786.1 JXJN01021770 KK107199 EZA55509.1 ACPB03016234 KZ288185 PBC34916.1 KK855619 PTY23302.1 GEDC01008695 JAS28603.1 AAZX01001195 CM000070 EIM52457.2 GL447755 EFN86021.1 KK852660 KDR19130.1 NNAY01004201 OXU18206.1 PNF21473.1 GDIQ01242201 GDIQ01210192 JAK09524.1 GDIQ01229038 JAK22687.1 GDIP01144919 GDIP01058394 LRGB01001361 JAJ78483.1 KZS12519.1
Proteomes
UP000007151
UP000009046
UP000053268
UP000008820
UP000092461
UP000069940
+ More
UP000249989 UP000092462 UP000000673 UP000075885 UP000092553 UP000069272 UP000075886 UP000075883 UP000075920 UP000075901 UP000053825 UP000075884 UP000075880 UP000076408 UP000030765 UP000075900 UP000075881 UP000076407 UP000075840 UP000075903 UP000007062 UP000002320 UP000075882 UP000007266 UP000235965 UP000076502 UP000095300 UP000002282 UP000008792 UP000075902 UP000095301 UP000009192 UP000007755 UP000037069 UP000008744 UP000092444 UP000001292 UP000000803 UP000000304 UP000268350 UP000008711 UP000001070 UP000007801 UP000007798 UP000078542 UP000192221 UP000078540 UP000000311 UP000183832 UP000078541 UP000075809 UP000005205 UP000053105 UP000092460 UP000053097 UP000092445 UP000015103 UP000078200 UP000242457 UP000005203 UP000091820 UP000002358 UP000001819 UP000008237 UP000027135 UP000215335 UP000076858
UP000249989 UP000092462 UP000000673 UP000075885 UP000092553 UP000069272 UP000075886 UP000075883 UP000075920 UP000075901 UP000053825 UP000075884 UP000075880 UP000076408 UP000030765 UP000075900 UP000075881 UP000076407 UP000075840 UP000075903 UP000007062 UP000002320 UP000075882 UP000007266 UP000235965 UP000076502 UP000095300 UP000002282 UP000008792 UP000075902 UP000095301 UP000009192 UP000007755 UP000037069 UP000008744 UP000092444 UP000001292 UP000000803 UP000000304 UP000268350 UP000008711 UP000001070 UP000007801 UP000007798 UP000078542 UP000192221 UP000078540 UP000000311 UP000183832 UP000078541 UP000075809 UP000005205 UP000053105 UP000092460 UP000053097 UP000092445 UP000015103 UP000078200 UP000242457 UP000005203 UP000091820 UP000002358 UP000001819 UP000008237 UP000027135 UP000215335 UP000076858
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A212EJ64
A0A2H1WTX1
E0VVB8
A0A194PJN3
A0A2W1BBJ5
A0A1S4G0S6
+ More
Q16HF5 A0A1B0CTV7 A0A2M4AAL0 A0A182G7Z6 A0A1B0DQ19 W5JAV9 A0A182PP74 A0A2M4BFU1 A0A0M4EVW7 A0A2M4BFL6 A0A182FAN7 A0A182QZ00 A0A182MQA6 A0A2M3Z490 A0A182W3G3 A0A182SPI1 A0A0L7QL47 A0A182NGF9 A0A2M3ZE23 A0A182JDJ3 A0A182Y8S1 A0A084WP56 A0A182RE39 A0A182K2Q5 A0A182XII5 A0A182HYP4 A0A182VMM3 Q7QA02 A0A1Y1L388 B0X1J7 A0A182KZJ3 D6X2I0 A0A2J7PYU2 A0A154P9T0 A0A0A1XEJ8 A0A0K8VNI7 A0A034VLB8 A0A1I8P161 A0A0R1E237 B4LXU7 A0A182TDM7 A0A1I8N0G7 B4K5C5 F4WUV5 W8B8E7 T1PQ56 B4PKL6 A0A0C9RT20 A0A0L0CJP2 B4G6D2 A0A1B0FN49 A0A1B6LC76 B4HJ91 Q9VH23 B4QVB1 A0A3B0KNI3 B3P4K8 B4JTX3 B3LWS7 B4NA47 A0A195CXL5 A0A1W4W076 A0A195ATX6 A0A1W4VZC8 E2AKF2 A0A1J1HK80 A0A151JZ33 A0A151WW69 A0A158P0U0 A0A336LTC4 A0A0M8ZW79 A0A069DVL9 A0A146M150 A0A0A9XRG9 A0A1B0BWG6 A0A026WK81 A0A1B0A7W4 T1HWC2 A0A1A9V8S2 A0A2A3EUM4 A0A2R7WUS0 A0A088AF18 A0A1B6DSH2 A0A1A9WNN2 K7IVC6 I5ANC2 E2BE34 A0A067R723 A0A232EIM5 A0A2J7PYQ9 A0A0P5I2A2 A0A0P5HBI7 A0A0P5FQZ2
Q16HF5 A0A1B0CTV7 A0A2M4AAL0 A0A182G7Z6 A0A1B0DQ19 W5JAV9 A0A182PP74 A0A2M4BFU1 A0A0M4EVW7 A0A2M4BFL6 A0A182FAN7 A0A182QZ00 A0A182MQA6 A0A2M3Z490 A0A182W3G3 A0A182SPI1 A0A0L7QL47 A0A182NGF9 A0A2M3ZE23 A0A182JDJ3 A0A182Y8S1 A0A084WP56 A0A182RE39 A0A182K2Q5 A0A182XII5 A0A182HYP4 A0A182VMM3 Q7QA02 A0A1Y1L388 B0X1J7 A0A182KZJ3 D6X2I0 A0A2J7PYU2 A0A154P9T0 A0A0A1XEJ8 A0A0K8VNI7 A0A034VLB8 A0A1I8P161 A0A0R1E237 B4LXU7 A0A182TDM7 A0A1I8N0G7 B4K5C5 F4WUV5 W8B8E7 T1PQ56 B4PKL6 A0A0C9RT20 A0A0L0CJP2 B4G6D2 A0A1B0FN49 A0A1B6LC76 B4HJ91 Q9VH23 B4QVB1 A0A3B0KNI3 B3P4K8 B4JTX3 B3LWS7 B4NA47 A0A195CXL5 A0A1W4W076 A0A195ATX6 A0A1W4VZC8 E2AKF2 A0A1J1HK80 A0A151JZ33 A0A151WW69 A0A158P0U0 A0A336LTC4 A0A0M8ZW79 A0A069DVL9 A0A146M150 A0A0A9XRG9 A0A1B0BWG6 A0A026WK81 A0A1B0A7W4 T1HWC2 A0A1A9V8S2 A0A2A3EUM4 A0A2R7WUS0 A0A088AF18 A0A1B6DSH2 A0A1A9WNN2 K7IVC6 I5ANC2 E2BE34 A0A067R723 A0A232EIM5 A0A2J7PYQ9 A0A0P5I2A2 A0A0P5HBI7 A0A0P5FQZ2
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.988393
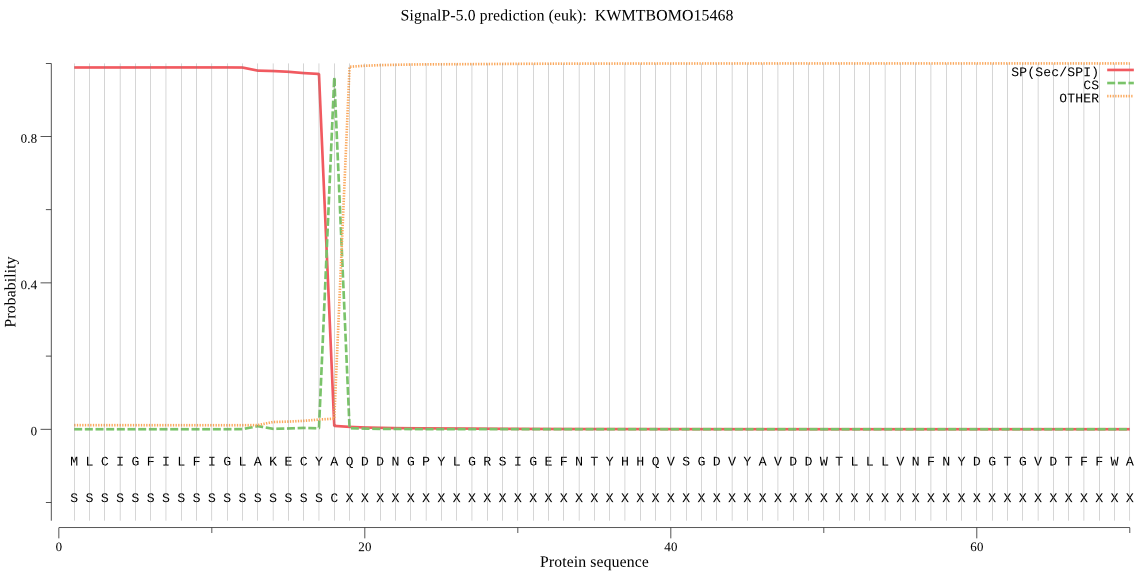
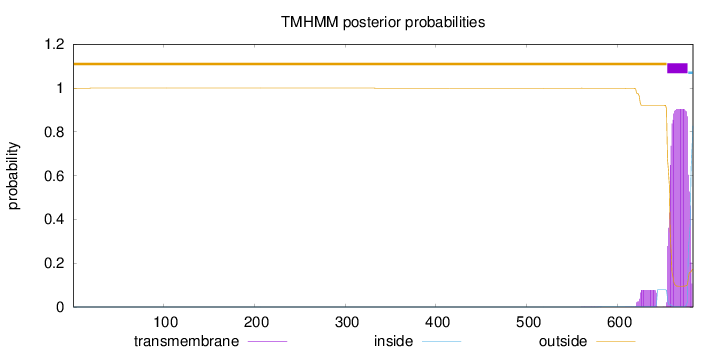
Length:
683
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.02951
Exp number, first 60 AAs:
0.00064
Total prob of N-in:
0.00040
outside
1 - 654
TMhelix
655 - 677
inside
678 - 683
Population Genetic Test Statistics
Pi
19.615476
Theta
23.193382
Tajima's D
-1.517079
CLR
181.864477
CSRT
0.0543972801359932
Interpretation
Uncertain
