Gene
KWMTBOMO15459
Pre Gene Modal
BGIBMGA004756
Annotation
PREDICTED:_lipase_member_H-B-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.955 PlasmaMembrane Reliability : 1.338
Sequence
CDS
ATGTGGAGGCCGGCGCTTTGCTGCCTCGCCGTGCTCCTGCCCAGCTTGTTGGCCGACCACTTCGACGAACTCCTCAGGACTGTCTCGCAGACCTGTGAAGTGCTTCCACTAAATATTCTCTTCTCGCGTCAAGTGACCCTCGAGTCTTCACACCTGGACGTAGTGGAATTCATGAAAGCCTCACGCCGAACGTATAAGCTTACCAAAGCTCATCTTAAAATCAGGCCCATGAAACTTAAAAGCCTGGTTCTCTACATCCCAGGGTGGTGGAACACGCCTAATGATGAGTCTTCTGTGGCTATAGTGAACTCTTTACTACAGAGGAATACTGTAGTATTTCTATTAGACACATGCCTCTCTTTCTGCAGAGGATACGTCGACTCAGCGTCACGAATCACAGCCATATCGAACTCTTTGTTCTCCTTCATGGAGAAGTTACACCAAGATGGATTTCCTATGGCTTCAGTTCATATTGTCGGCTTCAGTTTAGGTGCTCACGTGGCAGGCGCTGCTGGCAGATTGGTTCACTCCAAACTGTACAGTAAATTCGGTAAGATCACAGCTTTGGACCCCGCACGGCCATGCTTCGCGAGAACCGACAATAAACTTGAGAAAAATGATGCGTTATTCGTGCAAGTAATTCACGCCAGCACCGGGGTCCTGGGAGTCGAAGAGGCGACCGGCCATGTAGATGTCTACATCAACGGTCTGTCCGGGAAGCAGCCGGAATGTAGAAACATGAGTATCACCTTCGAATGCGATCACGCTCAAGCCTGGAAATTATATTCTGCGTCTGTTGTGAACGAAAATTCACTTACAGGCAGAAGATGTGAGAGCTGGCGGGAGTTAGAAAGAGAAAATTGCACTGGAGATGAGACTGTTCTTGGCTATGGCTGTCGATCGGATGTCAAAGGAGTTTTCTTGTACAAGTCGAAGGAAAGTAAAAAAAGGCAAGCGGTTAAATTGAAAGTGTTCAATCCTTTAGATTTTAGAACATGGTTTTCGAGATAG
Protein
MWRPALCCLAVLLPSLLADHFDELLRTVSQTCEVLPLNILFSRQVTLESSHLDVVEFMKASRRTYKLTKAHLKIRPMKLKSLVLYIPGWWNTPNDESSVAIVNSLLQRNTVVFLLDTCLSFCRGYVDSASRITAISNSLFSFMEKLHQDGFPMASVHIVGFSLGAHVAGAAGRLVHSKLYSKFGKITALDPARPCFARTDNKLEKNDALFVQVIHASTGVLGVEEATGHVDVYINGLSGKQPECRNMSITFECDHAQAWKLYSASVVNENSLTGRRCESWRELERENCTGDETVLGYGCRSDVKGVFLYKSKESKKRQAVKLKVFNPLDFRTWFSR
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9J5G6
A0A2H1VWW3
A0A2A4J3D6
A0A3S2LQ16
A0A212EW19
A0A0L7L2Y2
+ More
A0A194RK22 A0A2W1BWG2 A0A2H1W7F1 A0A0L7L8R4 A0A2H1W7Q1 A0A3S2NZZ7 A0A2H1W795 A0A1L8EFA4 A0A194PSE8 A0A1L8EFD1 A0A0L0C2D6 A0A194RQL7 H9JWY8 A0A2H1WNR8 A0A2H1X3Q9 A0A2A4JYV7 A0A2H1WL84 A0A2A4JGD9 A0A067QPW6 D6WXW0 A0A1L8E9J8 A0A1L8EB18 A0A220K8L9 U5EEG1 A0A1I8Q0V7 A0A1I8MA26 N6TY43 Q17N47 A0A1S4EX28 A0A1L8EAZ7 A0A2P8YQG3 A0A0L0CRF9 A0A336LE83 A0A2W1BXY3 A0A1L8E748 A0A0K8VXK1 A0A1I8MH84 A0A1W4VJJ8 A0A146LJU7 A0A0A9X8M8 A0A0K8SRI3 A0A1I8MJ22 A0A0K8U3C5 T1PDK2 A0A1J1HIX6 A0A034WKC4 A0A0A1WYA6 W8CB58 A0A1I8MR73 A0A146LFH2 A0A194QC74 E9IDK7 A0A0K8SS21 A0A1B0CTT6 W8BW27 A0A1B0D1A0 B4LGE8 A0A1A9V6P9 B4R6E4 A0A2A4J5Z0 A0A0A9XTP0 B4MEV6 B0X7X7 A0A1I8MK50 A0A1B0FNF4 A0A212FKA5 A0A182TWL0 A0A182GHZ7 B0XB54 A0A1I8MK53
A0A194RK22 A0A2W1BWG2 A0A2H1W7F1 A0A0L7L8R4 A0A2H1W7Q1 A0A3S2NZZ7 A0A2H1W795 A0A1L8EFA4 A0A194PSE8 A0A1L8EFD1 A0A0L0C2D6 A0A194RQL7 H9JWY8 A0A2H1WNR8 A0A2H1X3Q9 A0A2A4JYV7 A0A2H1WL84 A0A2A4JGD9 A0A067QPW6 D6WXW0 A0A1L8E9J8 A0A1L8EB18 A0A220K8L9 U5EEG1 A0A1I8Q0V7 A0A1I8MA26 N6TY43 Q17N47 A0A1S4EX28 A0A1L8EAZ7 A0A2P8YQG3 A0A0L0CRF9 A0A336LE83 A0A2W1BXY3 A0A1L8E748 A0A0K8VXK1 A0A1I8MH84 A0A1W4VJJ8 A0A146LJU7 A0A0A9X8M8 A0A0K8SRI3 A0A1I8MJ22 A0A0K8U3C5 T1PDK2 A0A1J1HIX6 A0A034WKC4 A0A0A1WYA6 W8CB58 A0A1I8MR73 A0A146LFH2 A0A194QC74 E9IDK7 A0A0K8SS21 A0A1B0CTT6 W8BW27 A0A1B0D1A0 B4LGE8 A0A1A9V6P9 B4R6E4 A0A2A4J5Z0 A0A0A9XTP0 B4MEV6 B0X7X7 A0A1I8MK50 A0A1B0FNF4 A0A212FKA5 A0A182TWL0 A0A182GHZ7 B0XB54 A0A1I8MK53
Pubmed
EMBL
BABH01028086
ODYU01004943
SOQ45331.1
NWSH01003696
PCG65913.1
RSAL01000642
+ More
RVE41276.1 AGBW02012106 OWR45657.1 JTDY01003257 KOB69863.1 KQ460118 KPJ17685.1 KZ149926 PZC77547.1 ODYU01006796 SOQ48977.1 JTDY01002277 KOB71750.1 SOQ48976.1 RSAL01000012 RVE53468.1 SOQ48975.1 GFDG01001409 JAV17390.1 KQ459601 KPI94050.1 GFDG01001408 JAV17391.1 JRES01000987 KNC26392.1 KQ459833 KPJ19782.1 BABH01041781 ODYU01009953 SOQ54715.1 ODYU01013233 SOQ59929.1 NWSH01000413 PCG76562.1 ODYU01009427 SOQ53845.1 NWSH01001670 PCG70462.1 KK853686 KDQ97822.1 KQ971362 EFA07925.2 GFDG01003510 JAV15289.1 GFDG01002871 JAV15928.1 MF319718 ASJ26432.1 GANO01004326 JAB55545.1 APGK01056924 APGK01056925 KB741277 KB632155 ENN71207.1 ERL89184.1 CH477202 EAT48096.1 GFDG01002872 JAV15927.1 PYGN01000432 PSN46485.1 JRES01000120 KNC34019.1 UFQS01003529 UFQT01003529 SSX15734.1 SSX35086.1 PZC77546.1 GFDG01004238 JAV14561.1 GDHF01008697 JAI43617.1 GDHC01011552 JAQ07077.1 GBHO01027613 JAG15991.1 GBRD01009842 JAG55982.1 GDHF01031494 JAI20820.1 KA646774 AFP61403.1 CVRI01000001 CRK86233.1 GAKP01004764 JAC54188.1 GBXI01010636 JAD03656.1 GAMC01000754 JAC05802.1 GDHC01012863 JAQ05766.1 KQ459193 KPJ03024.1 GL762454 EFZ21361.1 GBRD01009843 JAG55981.1 AJWK01027968 AJWK01027969 AJWK01027970 GAMC01009104 JAB97451.1 AJVK01002498 CH940647 EDW70477.1 CM000366 EDX18194.1 NWSH01003103 PCG66944.1 GBHO01020355 JAG23249.1 CH940664 EDW63081.1 DS232468 EDS42196.1 CCAG010000013 AGBW02008082 OWR54177.1 JXUM01065049 KQ562326 KXJ76141.1 DS232614 EDS44107.1
RVE41276.1 AGBW02012106 OWR45657.1 JTDY01003257 KOB69863.1 KQ460118 KPJ17685.1 KZ149926 PZC77547.1 ODYU01006796 SOQ48977.1 JTDY01002277 KOB71750.1 SOQ48976.1 RSAL01000012 RVE53468.1 SOQ48975.1 GFDG01001409 JAV17390.1 KQ459601 KPI94050.1 GFDG01001408 JAV17391.1 JRES01000987 KNC26392.1 KQ459833 KPJ19782.1 BABH01041781 ODYU01009953 SOQ54715.1 ODYU01013233 SOQ59929.1 NWSH01000413 PCG76562.1 ODYU01009427 SOQ53845.1 NWSH01001670 PCG70462.1 KK853686 KDQ97822.1 KQ971362 EFA07925.2 GFDG01003510 JAV15289.1 GFDG01002871 JAV15928.1 MF319718 ASJ26432.1 GANO01004326 JAB55545.1 APGK01056924 APGK01056925 KB741277 KB632155 ENN71207.1 ERL89184.1 CH477202 EAT48096.1 GFDG01002872 JAV15927.1 PYGN01000432 PSN46485.1 JRES01000120 KNC34019.1 UFQS01003529 UFQT01003529 SSX15734.1 SSX35086.1 PZC77546.1 GFDG01004238 JAV14561.1 GDHF01008697 JAI43617.1 GDHC01011552 JAQ07077.1 GBHO01027613 JAG15991.1 GBRD01009842 JAG55982.1 GDHF01031494 JAI20820.1 KA646774 AFP61403.1 CVRI01000001 CRK86233.1 GAKP01004764 JAC54188.1 GBXI01010636 JAD03656.1 GAMC01000754 JAC05802.1 GDHC01012863 JAQ05766.1 KQ459193 KPJ03024.1 GL762454 EFZ21361.1 GBRD01009843 JAG55981.1 AJWK01027968 AJWK01027969 AJWK01027970 GAMC01009104 JAB97451.1 AJVK01002498 CH940647 EDW70477.1 CM000366 EDX18194.1 NWSH01003103 PCG66944.1 GBHO01020355 JAG23249.1 CH940664 EDW63081.1 DS232468 EDS42196.1 CCAG010000013 AGBW02008082 OWR54177.1 JXUM01065049 KQ562326 KXJ76141.1 DS232614 EDS44107.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000037510
UP000053240
+ More
UP000053268 UP000037069 UP000027135 UP000007266 UP000095300 UP000095301 UP000019118 UP000030742 UP000008820 UP000245037 UP000192221 UP000183832 UP000092461 UP000092462 UP000008792 UP000078200 UP000000304 UP000002320 UP000092444 UP000075902 UP000069940 UP000249989
UP000053268 UP000037069 UP000027135 UP000007266 UP000095300 UP000095301 UP000019118 UP000030742 UP000008820 UP000245037 UP000192221 UP000183832 UP000092461 UP000092462 UP000008792 UP000078200 UP000000304 UP000002320 UP000092444 UP000075902 UP000069940 UP000249989
Interpro
Gene 3D
ProteinModelPortal
H9J5G6
A0A2H1VWW3
A0A2A4J3D6
A0A3S2LQ16
A0A212EW19
A0A0L7L2Y2
+ More
A0A194RK22 A0A2W1BWG2 A0A2H1W7F1 A0A0L7L8R4 A0A2H1W7Q1 A0A3S2NZZ7 A0A2H1W795 A0A1L8EFA4 A0A194PSE8 A0A1L8EFD1 A0A0L0C2D6 A0A194RQL7 H9JWY8 A0A2H1WNR8 A0A2H1X3Q9 A0A2A4JYV7 A0A2H1WL84 A0A2A4JGD9 A0A067QPW6 D6WXW0 A0A1L8E9J8 A0A1L8EB18 A0A220K8L9 U5EEG1 A0A1I8Q0V7 A0A1I8MA26 N6TY43 Q17N47 A0A1S4EX28 A0A1L8EAZ7 A0A2P8YQG3 A0A0L0CRF9 A0A336LE83 A0A2W1BXY3 A0A1L8E748 A0A0K8VXK1 A0A1I8MH84 A0A1W4VJJ8 A0A146LJU7 A0A0A9X8M8 A0A0K8SRI3 A0A1I8MJ22 A0A0K8U3C5 T1PDK2 A0A1J1HIX6 A0A034WKC4 A0A0A1WYA6 W8CB58 A0A1I8MR73 A0A146LFH2 A0A194QC74 E9IDK7 A0A0K8SS21 A0A1B0CTT6 W8BW27 A0A1B0D1A0 B4LGE8 A0A1A9V6P9 B4R6E4 A0A2A4J5Z0 A0A0A9XTP0 B4MEV6 B0X7X7 A0A1I8MK50 A0A1B0FNF4 A0A212FKA5 A0A182TWL0 A0A182GHZ7 B0XB54 A0A1I8MK53
A0A194RK22 A0A2W1BWG2 A0A2H1W7F1 A0A0L7L8R4 A0A2H1W7Q1 A0A3S2NZZ7 A0A2H1W795 A0A1L8EFA4 A0A194PSE8 A0A1L8EFD1 A0A0L0C2D6 A0A194RQL7 H9JWY8 A0A2H1WNR8 A0A2H1X3Q9 A0A2A4JYV7 A0A2H1WL84 A0A2A4JGD9 A0A067QPW6 D6WXW0 A0A1L8E9J8 A0A1L8EB18 A0A220K8L9 U5EEG1 A0A1I8Q0V7 A0A1I8MA26 N6TY43 Q17N47 A0A1S4EX28 A0A1L8EAZ7 A0A2P8YQG3 A0A0L0CRF9 A0A336LE83 A0A2W1BXY3 A0A1L8E748 A0A0K8VXK1 A0A1I8MH84 A0A1W4VJJ8 A0A146LJU7 A0A0A9X8M8 A0A0K8SRI3 A0A1I8MJ22 A0A0K8U3C5 T1PDK2 A0A1J1HIX6 A0A034WKC4 A0A0A1WYA6 W8CB58 A0A1I8MR73 A0A146LFH2 A0A194QC74 E9IDK7 A0A0K8SS21 A0A1B0CTT6 W8BW27 A0A1B0D1A0 B4LGE8 A0A1A9V6P9 B4R6E4 A0A2A4J5Z0 A0A0A9XTP0 B4MEV6 B0X7X7 A0A1I8MK50 A0A1B0FNF4 A0A212FKA5 A0A182TWL0 A0A182GHZ7 B0XB54 A0A1I8MK53
PDB
1W52
E-value=1.03475e-20,
Score=246
Ontologies
GO
PANTHER
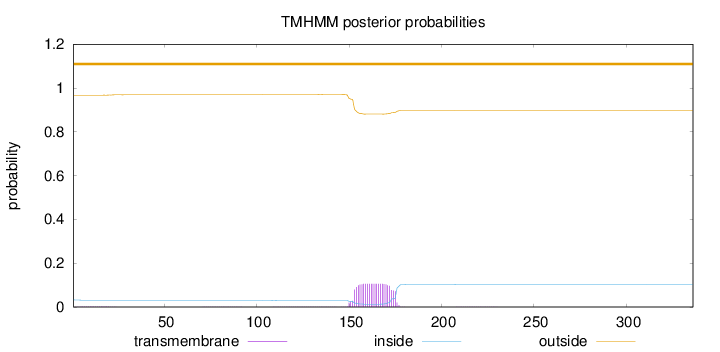
Topology
Subcellular location
Secreted
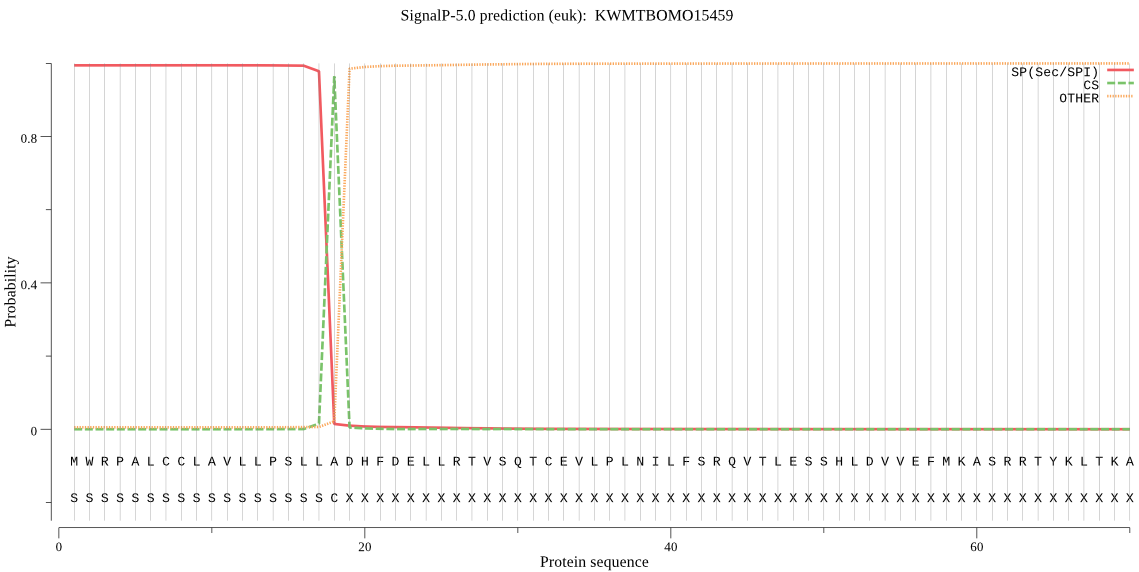
SignalP
Position: 1 - 18,
Likelihood: 0.994155
Length:
336
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.44052
Exp number, first 60 AAs:
0.03044
Total prob of N-in:
0.03261
outside
1 - 336
Population Genetic Test Statistics
Pi
181.043664
Theta
173.197576
Tajima's D
0.297502
CLR
0.229687
CSRT
0.457477126143693
Interpretation
Uncertain
