Pre Gene Modal
BGIBMGA004754
Annotation
PREDICTED:_nuclear_pore_complex_protein_Nup133_[Bombyx_mori]
Full name
Nuclear pore complex protein Nup133
Alternative Name
133 kDa nucleoporin
Location in the cell
Cytoplasmic Reliability : 1.531 PlasmaMembrane Reliability : 1.243
Sequence
CDS
ATGCAGTCACAAAGCGAACAAACTGGCGATGTGATCTATAAAACTCCATCGATGACACTAGAAACGTACGGGGCACCTATGCCAGTGATGGTCACAGAGGCTCTTGCGTTCGCAAGTGGAGACATCTCAGTGCGCCTCTCGACCTGTGGCTGGTGTTGGGTAGTGTCTGGTCGTCGCATCCTGGCTTGGGAGCAATCTCCAATAGATACTGATGAAGATCCAGCCAATGTGACTGGCATATCAGCTGCTCGAGAGCTCACATTACCTCAAACAGATTTAGCTCACAAGGCTGATTTAGTTGTACTGTTTTATGGAAATGACTCTCAGATGCCATCCTGTATTGGAGTTTCACCTGAGGGTGTGATTCGCTACTGGCCCTCAGTAGGACAAGAAGAGGTTTATGTAGATGTAACATGTGAACTAGCAGGTCAAGAGTGTGAGAAACTAGGAGATTACACCAGCTCTGGCCTCATACTGGCTACTACGACTTGTACCGTGGTGCTATTGACTCCTACTGTTCTTGATGGAAGAGCAACAGTGACTTGCTGCACTTTACGTCCACCGAGTGGTTGGCTAGGAGGTATAGGACGACGTGTATCACTGTTGTTCTTTGGATCTATGCCTGCACACACAGATACGCGGCTGGTGGGTGCGGTGTGGGCGGGGGACTGCGTGTGGCTGGTGGCTCGCGGGCCAACGCTGCAGGTGTGGCGCCCTCCCGCGCTGCTGCACCACCACTCGCTGCGGGGCCCGCTGGCGCTCGCCGCGCGCCCGCATCTGCAGCCTCACGGTGACTTGAACAGCTTGGAGATAATGGCTCTGGATGTGCAGGCCAGTGGGAACGATGGCTTCTTGCTACTAATCGCAGCAGTTAATGTAACGAGGTCACCGGAAATGAGATATGCTATTGCTCACATCAACGTAGAAGATCCATCGTCTCCTCGTGTAAGTTCATTTGTTCCTGTCCGGAATTTTCCTCAGGAAGCGGATGAGCCTCCGCGGCTTTTGCCCTTCGGAAACAAAGCGTTATTGTATACATCCAAATACATCGCTTTGATTTCACTAGCAGCCACGACGGACGCTGAAGTTGTGGAGGCGGCCAGTGGAGACCGCGTGCTCAGCGCCGCGCGCTCCCCGCACGGACCCGCGCTGTTCACCGCCGCGCACGGCCTGCTCGCACTGCGTCTGCACTCGCACGCCGCAGCTGCATGTGACAGTCCGATCGGCTCCTCGAGTCAAGCTGACATGTATGAAGGAAATTTATCGCTGTACGAAATTGATCCTTGCGAGATGAGCGCGGTGGTGACGGACGCGTGCGGGAAACTGAAGACGGCCTTCCTGTTCCACCTGCGGCGGGACTTCACCTCGTGCCGCGAGCTGCTGGACGAGCTGTTCCCGCCGCCCGGCCAGCACGCGCCGCAGGAGCTCGACGTGGACGCGCCCCTGGACCGCACCGTGCTCGCCGTGGCCACCGAGACGCTGGACGACATGCCCGCGGGGGATCCTAGGTGGAAGCGCTCGAGCGGACAGCCCACCCACATCCCGCTGGGCAGCTCCACGGCGCTGCAGATAGAAGCCCAGCTCCTGGACAAGCAGAGAGCGCTGGCACTCTTCTTCGACTTCCTGAGAGCGACCGGCCTGTGGCAGAGGTTGGGCCTCGTCACTGTAGAGGACGAGGGCGTGGCCAGCACCACGAGCGCGCTGTGCCAGGCCTGGGAGCGCGTGCGCGGCGCCGCGGCCCTGCGCCGCCTGCAGCGGGGGACCCACGCCGCCTTCATAGACTCCATTATATGTGAGGCGGTGCGCGGCTGGCAACACCGCGAGGAGGAGAGCGTGCACGCGGCGCTGTCCAGTGGCGCGCTGTCCCCGACGGACGCGTGCTACCGTCGCGTGTCCCGCCTGCCAGCCGTGCTGCTGATGCTCACCCGGCCCGTGCCGTCCGACCGCCCGGCGGCGCTTGCCGTGCTTAGTGGCGTGCTATCGGAGATGCTGAAGGCTCGCAAGTCGTGGCAGAACGTGTCGCCGCCCCCGCTGGGCTCCTCGGCGCTGCTCACCCCCGCGCTGCACCTCATCAGGAAGAGTCTCACTGAATTCGAGAACAAGGGGGGCGATGCGACGCTGCGCGCGCAGGTGTGCGAGGCGGCCGCGGGGCTGGCCGACGTGCTGCTGGCCGGCGCGCAGGCCTCCCCCGCCTACCCCGCGCTGCGCACCGACCTCATCCGCCCCTTCGTGGAGCAGGGGGAGCCGGAGCGCGCGGCGGCCCTGGCGGAGAAGTACCAGGAGTTCTCCGTGCTGGTGGAGCTGTGCGTGTCGGGCGGCGACACGGCGCGCCTGTGCGACTACGTGGACCGGTACAGCGACCGCGGTATGGCGGAGGTCGCGTACGAGTGGATGTCGCGCAAGGGTGGCGCCGCCGCCGCCGCCCTCGTCAGAGGCGCTGGGGGCGGAGCCCGGCTGACGGCGTGGCTGCGGCGGGAGCGCCCCGACCTGCTGTCGCTGCACCAGCTGCACCGCGGGGACCACGCCGCCGCCGCGCAAACCTTCAGCAGCCTCGCGCACGCTGAGACCGACAACCTCGAGCGGATGACGACGCTGGCGTCCCTGAGCAAGCTGTGCATGCTGGTGTCGGACGCGCCGGAGGAGGCCCCGGCGTGGGACGCCGTGCAGCGCCGCCTGGCCCTCGCCGAGCAGCACCGCGCCCTGCCCCGTGACGTCATGCCCAGTGACGTCACGCCGCAGCGCGTCATGCAGCCCCACGAGCTCGTCGAGGTGTACCTAAAAGCTGAGAACAAATCGTTGACTGAATACGACTACAAGAAGGCGCTGGACCTGACGGAGGGCGTGGAGGACCCCGAGCAGCGGCAGCGGCTGCGCCTGCGGGTGTGGTGCGCCAGCGTGCTGCGCGACGACTGGGCCGCCTGCAAGCCGGACGACCCCGCCGCCGAGCTCATGCAGAGGACCTTCTTCAAGCTCATAGACCTCGTCAACGTCATGGGCGACGACCTGGACGTGCTGCTGCCCGAGCTCCCCGAGCTGCTGTCGGCGCCGGAGCTGGCCGCGCTGGCTGCGGACTCCAAGTTCCACTACCTCCTCAAGTACGGGTACGAGTGCCTCCACCAGCGCACCGCGGACCAGAGCCCTGATGACTCTGCTTGA
Protein
MQSQSEQTGDVIYKTPSMTLETYGAPMPVMVTEALAFASGDISVRLSTCGWCWVVSGRRILAWEQSPIDTDEDPANVTGISAARELTLPQTDLAHKADLVVLFYGNDSQMPSCIGVSPEGVIRYWPSVGQEEVYVDVTCELAGQECEKLGDYTSSGLILATTTCTVVLLTPTVLDGRATVTCCTLRPPSGWLGGIGRRVSLLFFGSMPAHTDTRLVGAVWAGDCVWLVARGPTLQVWRPPALLHHHSLRGPLALAARPHLQPHGDLNSLEIMALDVQASGNDGFLLLIAAVNVTRSPEMRYAIAHINVEDPSSPRVSSFVPVRNFPQEADEPPRLLPFGNKALLYTSKYIALISLAATTDAEVVEAASGDRVLSAARSPHGPALFTAAHGLLALRLHSHAAAACDSPIGSSSQADMYEGNLSLYEIDPCEMSAVVTDACGKLKTAFLFHLRRDFTSCRELLDELFPPPGQHAPQELDVDAPLDRTVLAVATETLDDMPAGDPRWKRSSGQPTHIPLGSSTALQIEAQLLDKQRALALFFDFLRATGLWQRLGLVTVEDEGVASTTSALCQAWERVRGAAALRRLQRGTHAAFIDSIICEAVRGWQHREEESVHAALSSGALSPTDACYRRVSRLPAVLLMLTRPVPSDRPAALAVLSGVLSEMLKARKSWQNVSPPPLGSSALLTPALHLIRKSLTEFENKGGDATLRAQVCEAAAGLADVLLAGAQASPAYPALRTDLIRPFVEQGEPERAAALAEKYQEFSVLVELCVSGGDTARLCDYVDRYSDRGMAEVAYEWMSRKGGAAAAALVRGAGGGARLTAWLRRERPDLLSLHQLHRGDHAAAAQTFSSLAHAETDNLERMTTLASLSKLCMLVSDAPEEAPAWDAVQRRLALAEQHRALPRDVMPSDVTPQRVMQPHELVEVYLKAENKSLTEYDYKKALDLTEGVEDPEQRQRLRLRVWCASVLRDDWAACKPDDPAAELMQRTFFKLIDLVNVMGDDLDVLLPELPELLSAPELAALAADSKFHYLLKYGYECLHQRTADQSPDDSA
Summary
Description
Probable component of the nuclear pore complex (NPC) (Probable). Plays a role in NPC assembly and/or maintenance (PubMed:20547758).
Subunit
Forms part of the Nup107-Nup160 subcomplex in the nuclear pore.
Similarity
Belongs to the nucleoporin Nup133 family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. RsmB/NOP family.
Belongs to the class I-like SAM-binding methyltransferase superfamily. RsmB/NOP family.
Keywords
Complete proteome
mRNA transport
Nuclear pore complex
Nucleus
Protein transport
Reference proteome
Translocation
Transport
Feature
chain Nuclear pore complex protein Nup133
Uniprot
A0A3S2P298
A0A212EVZ1
A0A2J7RA04
E0VYF6
A0A1B0CE80
K7J5T7
+ More
A0A088A0Z5 A0A0L7QUQ5 A0A232EZI1 A0A2A3EN61 A0A0C9R9D3 A0A0M8ZYZ0 A0A1B6E0K1 E2BNB0 V4AAF7 U4U868 F4WA61 A0A158NHL0 A0A1I8NVE6 A0A1I8NVD9 A0A1I8NBJ4 A0A0A1WDU8 A1YK82 A0A182YLM2 A1YK81 A1YK79 A1YK83 A1YK85 A1YK77 Q9VCW3 A1YK80 A1YK78 A1YK86 E9FWZ0 A0A0P5MHN3 A0A0P5LZ98 A0A0P4XQF8 A0A0P5QA91 B4HER9 A1YK95 A1YK92 A1YK88 A1YK98 A1YK96 A0A1A9YIY0 A1YK94 A1YK97 A1YKA0 A1YK91 B4KBB3 A1YK89 B4PMD6 A1YK90 B4JRZ7 A0A1A9VQY3 A0A0M4F7F2 A0A0J7KD42 A0A0K8WAZ8 A0A0P5YWY4 B4M641 A0A131YL41 A0A2T7PFZ6 A0A2C9JQH2 A0A336M2M2 A0A293M9N9 A0A224YDA6 A0A034VG90 A0A0P6I4Z1 A0A0P5CMP7 A0A0P5VQ99 A0A0P6C8Q9 A0A0N8DD69 A0A0B7AV36 A0A0P5JC23 A0A0P5AEK7 A0A0P5JAB7 A0A0P5MDD3 A0A0P5IYI0 A0A0P5J5G0 A0A195ETN2 A0A195AUP7 G1K922 A0A151M2P4 A0A3B3S8N1
A0A088A0Z5 A0A0L7QUQ5 A0A232EZI1 A0A2A3EN61 A0A0C9R9D3 A0A0M8ZYZ0 A0A1B6E0K1 E2BNB0 V4AAF7 U4U868 F4WA61 A0A158NHL0 A0A1I8NVE6 A0A1I8NVD9 A0A1I8NBJ4 A0A0A1WDU8 A1YK82 A0A182YLM2 A1YK81 A1YK79 A1YK83 A1YK85 A1YK77 Q9VCW3 A1YK80 A1YK78 A1YK86 E9FWZ0 A0A0P5MHN3 A0A0P5LZ98 A0A0P4XQF8 A0A0P5QA91 B4HER9 A1YK95 A1YK92 A1YK88 A1YK98 A1YK96 A0A1A9YIY0 A1YK94 A1YK97 A1YKA0 A1YK91 B4KBB3 A1YK89 B4PMD6 A1YK90 B4JRZ7 A0A1A9VQY3 A0A0M4F7F2 A0A0J7KD42 A0A0K8WAZ8 A0A0P5YWY4 B4M641 A0A131YL41 A0A2T7PFZ6 A0A2C9JQH2 A0A336M2M2 A0A293M9N9 A0A224YDA6 A0A034VG90 A0A0P6I4Z1 A0A0P5CMP7 A0A0P5VQ99 A0A0P6C8Q9 A0A0N8DD69 A0A0B7AV36 A0A0P5JC23 A0A0P5AEK7 A0A0P5JAB7 A0A0P5MDD3 A0A0P5IYI0 A0A0P5J5G0 A0A195ETN2 A0A195AUP7 G1K922 A0A151M2P4 A0A3B3S8N1
Pubmed
EMBL
RSAL01000001
RVE55286.1
AGBW02012106
OWR45653.1
NEVH01006580
PNF37667.1
+ More
DS235845 EEB18412.1 AJWK01008657 AAZX01003923 KQ414735 KOC62309.1 NNAY01001572 OXU23558.1 KZ288215 PBC32579.1 GBYB01013014 JAG82781.1 KQ435821 KOX72473.1 GEDC01005859 JAS31439.1 GL449385 EFN82872.1 KB202163 ESO92060.1 KB631792 ERL86140.1 GL888041 EGI68917.1 ADTU01015920 GBXI01017113 JAC97178.1 EF058005 EF058007 ABL84940.1 ABL84942.1 EF058004 ABL84939.1 EF058002 ABL84937.1 EF058006 ABL84941.1 EF058008 ABL84943.1 EF058000 ABL84935.1 AE014297 BT003226 AY051721 AAK93145.1 AAO24981.1 EF058003 EF058010 ABL84938.1 ABL84945.1 EF058001 ABL84936.1 EF058009 ABL84944.1 GL732526 EFX88351.1 GDIQ01158058 GDIQ01034998 JAK93667.1 GDIQ01173640 JAK78085.1 GDIP01240567 JAI82834.1 GDIQ01123022 JAL28704.1 CH480815 EDW43230.1 EF058018 ABL84953.1 EF058015 ABL84950.1 EF058011 ABL84946.1 EF058021 ABL84956.1 EF058019 ABL84954.1 EF058017 ABL84952.1 EF058020 EF058022 ABL84955.1 ABL84957.1 EF058023 ABL84958.1 EF058014 ABL84949.1 CH933806 EDW16841.1 EF058012 EF058016 ABL84947.1 ABL84951.1 CM000160 EDW98041.1 EF058013 ABL84948.1 CH916373 EDV94537.1 CP012526 ALC47782.1 LBMM01009259 KMQ88267.1 GDHF01003951 JAI48363.1 GDIP01052539 LRGB01001361 JAM51176.1 KZS12347.1 CH940652 EDW59117.1 GEDV01009926 JAP78631.1 PZQS01000004 PVD32345.1 UFQT01000114 SSX20218.1 GFWV01012337 MAA37066.1 GFPF01004542 MAA15688.1 GAKP01018172 JAC40780.1 GDIQ01009414 JAN85323.1 GDIP01168309 JAJ55093.1 GDIP01096929 JAM06786.1 GDIP01019209 JAM84506.1 GDIP01045295 JAM58420.1 HACG01037873 CEK84738.1 GDIQ01227180 JAK24545.1 GDIP01213171 JAJ10231.1 GDIQ01204784 JAK46941.1 GDIQ01165552 JAK86173.1 GDIQ01207645 JAK44080.1 GDIQ01206289 JAK45436.1 KQ981975 KYN31598.1 KQ976737 KYM75963.1 AKHW03006780 KYO18784.1
DS235845 EEB18412.1 AJWK01008657 AAZX01003923 KQ414735 KOC62309.1 NNAY01001572 OXU23558.1 KZ288215 PBC32579.1 GBYB01013014 JAG82781.1 KQ435821 KOX72473.1 GEDC01005859 JAS31439.1 GL449385 EFN82872.1 KB202163 ESO92060.1 KB631792 ERL86140.1 GL888041 EGI68917.1 ADTU01015920 GBXI01017113 JAC97178.1 EF058005 EF058007 ABL84940.1 ABL84942.1 EF058004 ABL84939.1 EF058002 ABL84937.1 EF058006 ABL84941.1 EF058008 ABL84943.1 EF058000 ABL84935.1 AE014297 BT003226 AY051721 AAK93145.1 AAO24981.1 EF058003 EF058010 ABL84938.1 ABL84945.1 EF058001 ABL84936.1 EF058009 ABL84944.1 GL732526 EFX88351.1 GDIQ01158058 GDIQ01034998 JAK93667.1 GDIQ01173640 JAK78085.1 GDIP01240567 JAI82834.1 GDIQ01123022 JAL28704.1 CH480815 EDW43230.1 EF058018 ABL84953.1 EF058015 ABL84950.1 EF058011 ABL84946.1 EF058021 ABL84956.1 EF058019 ABL84954.1 EF058017 ABL84952.1 EF058020 EF058022 ABL84955.1 ABL84957.1 EF058023 ABL84958.1 EF058014 ABL84949.1 CH933806 EDW16841.1 EF058012 EF058016 ABL84947.1 ABL84951.1 CM000160 EDW98041.1 EF058013 ABL84948.1 CH916373 EDV94537.1 CP012526 ALC47782.1 LBMM01009259 KMQ88267.1 GDHF01003951 JAI48363.1 GDIP01052539 LRGB01001361 JAM51176.1 KZS12347.1 CH940652 EDW59117.1 GEDV01009926 JAP78631.1 PZQS01000004 PVD32345.1 UFQT01000114 SSX20218.1 GFWV01012337 MAA37066.1 GFPF01004542 MAA15688.1 GAKP01018172 JAC40780.1 GDIQ01009414 JAN85323.1 GDIP01168309 JAJ55093.1 GDIP01096929 JAM06786.1 GDIP01019209 JAM84506.1 GDIP01045295 JAM58420.1 HACG01037873 CEK84738.1 GDIQ01227180 JAK24545.1 GDIP01213171 JAJ10231.1 GDIQ01204784 JAK46941.1 GDIQ01165552 JAK86173.1 GDIQ01207645 JAK44080.1 GDIQ01206289 JAK45436.1 KQ981975 KYN31598.1 KQ976737 KYM75963.1 AKHW03006780 KYO18784.1
Proteomes
UP000283053
UP000007151
UP000235965
UP000009046
UP000092461
UP000002358
+ More
UP000005203 UP000053825 UP000215335 UP000242457 UP000053105 UP000008237 UP000030746 UP000030742 UP000007755 UP000005205 UP000095300 UP000095301 UP000076408 UP000000803 UP000000305 UP000001292 UP000092443 UP000009192 UP000002282 UP000001070 UP000078200 UP000092553 UP000036403 UP000076858 UP000008792 UP000245119 UP000076420 UP000078541 UP000078540 UP000001646 UP000050525 UP000261540
UP000005203 UP000053825 UP000215335 UP000242457 UP000053105 UP000008237 UP000030746 UP000030742 UP000007755 UP000005205 UP000095300 UP000095301 UP000076408 UP000000803 UP000000305 UP000001292 UP000092443 UP000009192 UP000002282 UP000001070 UP000078200 UP000092553 UP000036403 UP000076858 UP000008792 UP000245119 UP000076420 UP000078541 UP000078540 UP000001646 UP000050525 UP000261540
Interpro
SUPFAM
SSF53335
SSF53335
Gene 3D
ProteinModelPortal
A0A3S2P298
A0A212EVZ1
A0A2J7RA04
E0VYF6
A0A1B0CE80
K7J5T7
+ More
A0A088A0Z5 A0A0L7QUQ5 A0A232EZI1 A0A2A3EN61 A0A0C9R9D3 A0A0M8ZYZ0 A0A1B6E0K1 E2BNB0 V4AAF7 U4U868 F4WA61 A0A158NHL0 A0A1I8NVE6 A0A1I8NVD9 A0A1I8NBJ4 A0A0A1WDU8 A1YK82 A0A182YLM2 A1YK81 A1YK79 A1YK83 A1YK85 A1YK77 Q9VCW3 A1YK80 A1YK78 A1YK86 E9FWZ0 A0A0P5MHN3 A0A0P5LZ98 A0A0P4XQF8 A0A0P5QA91 B4HER9 A1YK95 A1YK92 A1YK88 A1YK98 A1YK96 A0A1A9YIY0 A1YK94 A1YK97 A1YKA0 A1YK91 B4KBB3 A1YK89 B4PMD6 A1YK90 B4JRZ7 A0A1A9VQY3 A0A0M4F7F2 A0A0J7KD42 A0A0K8WAZ8 A0A0P5YWY4 B4M641 A0A131YL41 A0A2T7PFZ6 A0A2C9JQH2 A0A336M2M2 A0A293M9N9 A0A224YDA6 A0A034VG90 A0A0P6I4Z1 A0A0P5CMP7 A0A0P5VQ99 A0A0P6C8Q9 A0A0N8DD69 A0A0B7AV36 A0A0P5JC23 A0A0P5AEK7 A0A0P5JAB7 A0A0P5MDD3 A0A0P5IYI0 A0A0P5J5G0 A0A195ETN2 A0A195AUP7 G1K922 A0A151M2P4 A0A3B3S8N1
A0A088A0Z5 A0A0L7QUQ5 A0A232EZI1 A0A2A3EN61 A0A0C9R9D3 A0A0M8ZYZ0 A0A1B6E0K1 E2BNB0 V4AAF7 U4U868 F4WA61 A0A158NHL0 A0A1I8NVE6 A0A1I8NVD9 A0A1I8NBJ4 A0A0A1WDU8 A1YK82 A0A182YLM2 A1YK81 A1YK79 A1YK83 A1YK85 A1YK77 Q9VCW3 A1YK80 A1YK78 A1YK86 E9FWZ0 A0A0P5MHN3 A0A0P5LZ98 A0A0P4XQF8 A0A0P5QA91 B4HER9 A1YK95 A1YK92 A1YK88 A1YK98 A1YK96 A0A1A9YIY0 A1YK94 A1YK97 A1YKA0 A1YK91 B4KBB3 A1YK89 B4PMD6 A1YK90 B4JRZ7 A0A1A9VQY3 A0A0M4F7F2 A0A0J7KD42 A0A0K8WAZ8 A0A0P5YWY4 B4M641 A0A131YL41 A0A2T7PFZ6 A0A2C9JQH2 A0A336M2M2 A0A293M9N9 A0A224YDA6 A0A034VG90 A0A0P6I4Z1 A0A0P5CMP7 A0A0P5VQ99 A0A0P6C8Q9 A0A0N8DD69 A0A0B7AV36 A0A0P5JC23 A0A0P5AEK7 A0A0P5JAB7 A0A0P5MDD3 A0A0P5IYI0 A0A0P5J5G0 A0A195ETN2 A0A195AUP7 G1K922 A0A151M2P4 A0A3B3S8N1
PDB
3I4R
E-value=1.71517e-20,
Score=249
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Located on both the cytoplasmic and nuclear sides of the nuclear pore. With evidence from 1 publications.
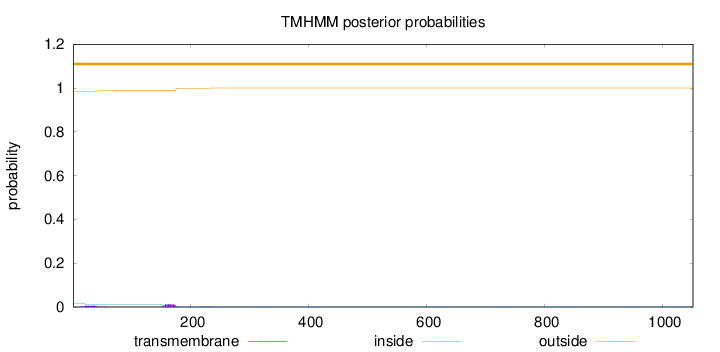
Length:
1051
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.386760000000001
Exp number, first 60 AAs:
0.12676
Total prob of N-in:
0.01718
outside
1 - 1051
Population Genetic Test Statistics
Pi
172.952746
Theta
18.993003
Tajima's D
-1.110189
CLR
1.454577
CSRT
0.116994150292485
Interpretation
Uncertain
