Gene
KWMTBOMO15455
Pre Gene Modal
BGIBMGA004765
Annotation
PREDICTED:_structural_maintenance_of_chromosomes_protein_2_[Amyelois_transitella]
Full name
Structural maintenance of chromosomes protein
Location in the cell
Cytoplasmic Reliability : 1.98 Nuclear Reliability : 1.543
Sequence
CDS
ATGTTTATTAAGTCAATAGTGTTAGATGGCTTCAAGTCTTACGGAAACCGAGTGGAAATTACTGGATTTGATCCAGAATTTAATGCGATCACTGGTCTAAATGGGACTGGAAAATCAAATATCTTAGATTCTATATGTTTCGTACTTGGAATCACAAATTTATCGAATGTACGAGCAAGCAGCTTGCAGGAGCTTATTTACAAGCATGGGCAGGCAGGTATCACCAAAGCAACAGTTAGCATTACCTTCGACAACAGAGATAAGAGACAATGTCCGATTGGCTATGAGAATCATGATGAGATTACTGTCACTAGACAGGTGGTGATGGGTGGTAAGAATAAATACCTTATAAATGGTATCAATGTCCAAAACAAGAGGGTCAGTGACTTATTCTGTTCGGTTCAGCTTAATGTCAATAACCCACACTTCCTCATCATGCAGGGAAGGATTACTAAGGTGCTAAATATGAAACCTCCTGAGATATTATCAATGGTAGAGGAAGCGGCAGGTACAAGAATGTATGAAGCTAAAAAACAAGCAGCACAGAAAACTATTGAGAAGAAAGATGCAAAGCTCCGGGAACTTAATGATATAATAAGAGAAGACATAGCACCTAAACTTCAGAAGCTGCAAGATGAAAGATCTCAGTTCCAAGAATATCAGAAGGTTGTCAGAGAATTGGAGAATATCACCAGGCTGTGTGTCGCTTGGAAGTATGTTTCTGCTGAAGAGCAAACCAAAGCAGCAGAGGGAAAAGTCACGCAAGTGCAGGACGACATTAAATTGAAAAAAGAGACTATTAAAAACAATGAGAAAGAAATGAAAGAGCTGGATAAGCAAGTTGTTGACTTAAACAAAAAACTGGATGAGGAGAGTGGGAATGTCCTCAAAGATTTGGAAGTAGAACTTCAAAACTTGGAGAAGGTTGAAGCAGCAGCAAGTTCACAATATAAAGGCAGCAAAGAAGCCCTGAGCGGACACGAGAAGAGAGTCAAACTACTCGAAAAGGCACTGGCTGAGGACGAGCAGGCTCTCACATCAAAGCAACAGATTCTTAATGAGGTCTCGTCAACATTCGAGAACCTTCGCACATCTAGTGAGACCAGTGAGAAAGCTTTAGCGGAAGCACAACAGAAGTTCTTGGCCGTGAGCACCGGGAAGGAAGATGCTTCTGAATCGCTGCAGGACCAGCTAATGGCTGCCAAACAACAGGCGTCGGAGGCGTCCACTCGCATCACCTCGGCCATGATGGACAAGAAGCACGCCGAGGAGAGATTGAAAACTTTGGAGAAGGAGTTCACTACCAGCAGCAAACAGTACGACGCGGATCAGGAGAGCATCCGGCGGCAGCAGGCGCAGGTCGAGCAGCTGGAGCGCGAGCTGGCGGGGCTGGCGCACGACGAGTCCCGCGTGACGTCACTGGGGGCGGCGGTGCGGGGCGCGCAGGGGGCGGCGCGGGGGGCGCGGGACCGGCGGGACGCGCTGGCGGCGGCGCTGCAGCGGTGCCACTTCCGGTACTCCCCCCCCGACCCGCACTTCGACCGCGCCAGGGTCGCAGGTACCGTCTGCAAGCTGATCGACGTGTGTGATCCGGTGTACTGCACCGCCCTGGAGACGGCGGCCGGAGGACGGTTATACAATGTAGTGGTGGACACCGAGGTCACGAGCAAGCTGCTGCTACAGCGGGGCAACCTGCAGACCAGAACTACCATCATACCGCTCAACAAGATCTCCGGACAGGTCATCGACAAGAGAATTGTGCAGATAGCTCAGGAGATTGGTGGAGGTCCGGAGAACGTGCAGCTGGCCTTGGACCTGATATCGTTCGACAGCCGCCTGCGCCCCGCCATGGCGTGGGTGTTCGGCTCCACGCTCGTGTGCCGCGACCTCGACACCGCCAAGCGCGTCACCTTCCACCGCGCCGTGCGCCGCCGCTGCGTCACGCTCGACGGCGACGTGTTCGACCCCTCCGGCACGCTCTCCGGCGGCGCCGCGCTCAAGGCACGACGACATAATATATCTTGTTTATATTTTTGTATTTTAGGCGGGTCGGTGCTGCTGCAGCTGCAGGAGCTGAAGCAGCTGGAGCAGCAGGTGCAGGCGGCCGAGGCGCGGCTGGCGCAGCTCACGGAGGAGCTGGGCTGCGCGCAGCAGGCTGCCGAGAGCTACGCCGCTATACAACAGAAGTGCGAGGCGGCGCGGCTGGGCGCGCGCGCGGCGGCGGCGCGGCTGGGCGGCACGGCGCGCGCGCAGCTGCACGCCGAGATCCAGGCTCTGCGCGCGCAGGCGGCCCAGCTCGCGGCGGCGCTGGAGCAGGACGAGCTCGTGCAGGACGAAGCGGCCGGCCGGGCCCGCGACCTGGAGGCCAAGCTCAAGGACATCAAGGGACACCGAGAGAGAGAATATAAGAAAGCAGAAGAGGCATTGAAGAAAGCAAAGCGAGACGTGGAGGTGCACGGCGGCGCGTGGAAGCTGCGCGAGCAGGAGCTGGAGACCCTGCGGCTGGAGGCGGCGGAGCTGCAGCGCGGCGCCGCGGCCGCACGCCACCAGCTGCACGAGCACGCGCGCGCCGCCGCCGACCTGGCACGCGCACTGCACGACGCCGCGCGCAGCCGGGACCACGCGCAGGAAGCAGTGAAGGAGATGCAGGCGCGAATAAAACATCAGAAAACGGAGATCGCGGGCCGGAGCACCGACATATCTCGCCTCGTGCAGCAGAAGGAGCGCCTGGCCAAGAGCAGCAGCGAGCTGGAGCTCGGCGTCAAGGAGCTGGACTACCGCATCAAGGAGCTGCAGACTGAGGCTCTCGATTGTCAGAAGAAGATGAGGCAACTGGAGTCGGAGAACCCGTGGATCCCGTCGGAGCGGCAGTACTTCGGCGCGCCGGGCGGCCTCTACGACTTCGAGGGTCGGCAGGCGCACGTGGCCGCCAGCCGCCTGCACCAGCTCGCCGAGCGCAGGGACCGCCTCGCGCGAGGACTCAACGCCAGGGCGCACACGCTGCTCGGCAAGGAGGAGGACCAGTACCAAGACGTCATGAGGAAGAAGCAGATCGTGGAGGCGGACAGAGCCAAGCTGGTGCAGGTCATGGCGGAGCTGGACGAGAAGAAGAAGAAGACGTTAGTGACGGCCTGCGAGCAAGTCAACCGGGACTTCGGGTCCATCTTCAGCACCCTGCTGCCCGGCGCGCAGGCCAAGCTGCAGCCGCCCGACGGACTCTCCGTGCTGGACGGGCTCGAGGTGAAGGTGGGGTTCAACATGACGTGGAAGGAGTCCCTGGGCGAGCTGTCGGGTGGGCAGCGGTCGCTGGTGGCGCTAGCGCTGGTGCTGGCGATGCTGCTGTTCAAGCCGGCTCCGCTGTACATCCTGGACGAGGTGGACGCGGCGCTGGACCTGTCGCACACGCAGAACATCGGCATCATGCTCAAGGAACACTTCCGACACTCGCAGTTCATCATCGTGTCGCTGAAGGACGGCATGTTCAACAACGCCAACGTTCTGTTCCGCACCCGCTTCGTGGACGGCATGTCCTCCGTGCAGCGCACCGCCGCGCGACGCTAG
Protein
MFIKSIVLDGFKSYGNRVEITGFDPEFNAITGLNGTGKSNILDSICFVLGITNLSNVRASSLQELIYKHGQAGITKATVSITFDNRDKRQCPIGYENHDEITVTRQVVMGGKNKYLINGINVQNKRVSDLFCSVQLNVNNPHFLIMQGRITKVLNMKPPEILSMVEEAAGTRMYEAKKQAAQKTIEKKDAKLRELNDIIREDIAPKLQKLQDERSQFQEYQKVVRELENITRLCVAWKYVSAEEQTKAAEGKVTQVQDDIKLKKETIKNNEKEMKELDKQVVDLNKKLDEESGNVLKDLEVELQNLEKVEAAASSQYKGSKEALSGHEKRVKLLEKALAEDEQALTSKQQILNEVSSTFENLRTSSETSEKALAEAQQKFLAVSTGKEDASESLQDQLMAAKQQASEASTRITSAMMDKKHAEERLKTLEKEFTTSSKQYDADQESIRRQQAQVEQLERELAGLAHDESRVTSLGAAVRGAQGAARGARDRRDALAAALQRCHFRYSPPDPHFDRARVAGTVCKLIDVCDPVYCTALETAAGGRLYNVVVDTEVTSKLLLQRGNLQTRTTIIPLNKISGQVIDKRIVQIAQEIGGGPENVQLALDLISFDSRLRPAMAWVFGSTLVCRDLDTAKRVTFHRAVRRRCVTLDGDVFDPSGTLSGGAALKARRHNISCLYFCILGGSVLLQLQELKQLEQQVQAAEARLAQLTEELGCAQQAAESYAAIQQKCEAARLGARAAAARLGGTARAQLHAEIQALRAQAAQLAAALEQDELVQDEAAGRARDLEAKLKDIKGHREREYKKAEEALKKAKRDVEVHGGAWKLREQELETLRLEAAELQRGAAAARHQLHEHARAAADLARALHDAARSRDHAQEAVKEMQARIKHQKTEIAGRSTDISRLVQQKERLAKSSSELELGVKELDYRIKELQTEALDCQKKMRQLESENPWIPSERQYFGAPGGLYDFEGRQAHVAASRLHQLAERRDRLARGLNARAHTLLGKEEDQYQDVMRKKQIVEADRAKLVQVMAELDEKKKKTLVTACEQVNRDFGSIFSTLLPGAQAKLQPPDGLSVLDGLEVKVGFNMTWKESLGELSGGQRSLVALALVLAMLLFKPAPLYILDEVDAALDLSHTQNIGIMLKEHFRHSQFIIVSLKDGMFNNANVLFRTRFVDGMSSVQRTAARR
Summary
Similarity
Belongs to the SMC family.
Uniprot
EMBL
Proteomes
Interpro
ProteinModelPortal
PDB
3L51
E-value=3.71168e-39,
Score=410
Ontologies
GO
Topology
Subcellular location
Nucleus
Length:
1186
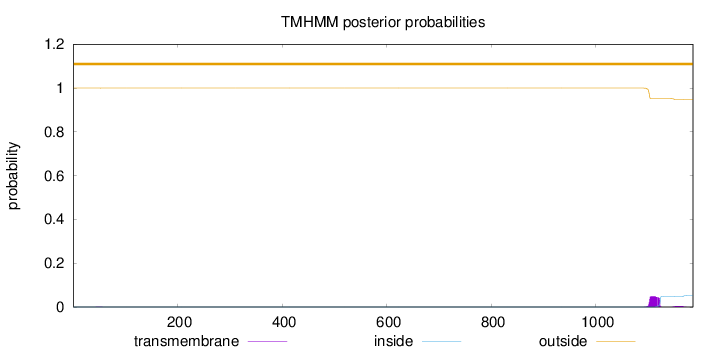
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.08904
Exp number, first 60 AAs:
0.00265
Total prob of N-in:
0.00016
outside
1 - 1186
Population Genetic Test Statistics
Pi
191.825512
Theta
169.578135
Tajima's D
0.158238
CLR
0.760526
CSRT
0.409529523523824
Interpretation
Uncertain
