Gene
KWMTBOMO15451
Pre Gene Modal
BGIBMGA004767
Annotation
PREDICTED:_nephrin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.791
Sequence
CDS
ATGGACGCCAAGACCAGAAGCCACACTCGACTACTAGAGCCCTACAACGAGGGAGATACCCTGGAACTTATTTGCGAAACATATGGTGGTGATCCAAGGCCCAGTGTGACTTGGTACTTGGAGAACACCGTCATCGACGACTCATTTGAGCAGCGCCCTGACGGAGTGACTGTCAACACCCTGACCTTCCCCAGTGTTGGTCGTCAGCATCTCAACGCAAGGCTCGTCTGTCAAGCTTCGAACACAAATCTAGCGCCACCGGAAACTAAGCTTTTAATACTTGATATAAACTTAAGGCCACTCAGCGTGCAGATCTTGAATAAGAGGCCGGAGCTTTCCGCAGACAGGACTTATGAACTTGAGTGCAAGACTACTGGATCCCGGCCCGAAGCTCAAGTTACGTGGTGGAAAGGTGGAAGACAATACAAAAGGAGTGCAGCCAAAAATTTCTCCGAGAGCAACGCAACGACCAGCGTCCTAACGTTTGTGCCTGTCGCCGAGGACCACGAGAAGACCCTGGTCTGTCGCGCCGAGAACCCCAGCGTGGTGGAATCATCGATAGAAGACAAATGGAAAATGAACGTGCATTATAACCCATCAAGATTAAACTTAGAAAAAAACTACTGA
Protein
MDAKTRSHTRLLEPYNEGDTLELICETYGGDPRPSVTWYLENTVIDDSFEQRPDGVTVNTLTFPSVGRQHLNARLVCQASNTNLAPPETKLLILDINLRPLSVQILNKRPELSADRTYELECKTTGSRPEAQVTWWKGGRQYKRSAAKNFSESNATTSVLTFVPVAEDHEKTLVCRAENPSVVESSIEDKWKMNVHYNPSRLNLEKNY
Summary
Uniprot
A0A194PPP2
A0A212EM07
H9J5G2
H9J5H7
A0A2A4JDY9
A0A212FI98
+ More
A0A194RK28 A0A194PPB3 A0A2H1WJ89 A0A2A4JRL1 A0A3S2MB94 A0A1B0CF37 A0A0A9ZBH5 A0A0K8T359 A0A1Y1NKS5 D6WPK9 A0A182UG36 A0A1W4WQT7 A0A1W4WRT9 A0A1W4WFY8 A0A1J1HQ06 A0A182LRH8 A0A182KC68 A0A182R5H8 A0A182N5E5 A0A2J7PYK2 A0A194RJS5 A0A067RCL4 A0A1B0BUJ4 N6U7Y1 A0A182X9U7 A0A154P4D6 A0A182L937 A0A182Y570 A0A1Y9IRW5 W5JRL3 A0A182HKS9 A0A182VRA0 A0A182P2F8 A0A182QRL9 A0A182IS29 A0A336MBR2 U4TXN3 A0A2J7PYN3 B0WA44 A0A1B0GLI9 A0A336M9R3 A0A1B6C6V9 A0A0P9ANK1 B4NKE4 A0A1I8PBW5 Q16ZD1 A0A1I8NIL5 A0A0L0BY62 Q7PXI0 A0A0Q9W1D8 A0A1S4GCH1 B4M6A4 A0A0M5JCQ5 A0A1A9UZ31 A0A1W7R8R5 B3LX91 A0A0R3NN42 A0A0A1WVT0 A0A034W9N8 A0A0K8U830 A0A1W4UR97 A0A0K8WMB4 B4G2W0 C0PTV4 W8B9C8 Q297Q6 B4KAA3 A0A1W4V371 A0A3B0KV58 A0A1W4V3H8 A0A1A9Z0I0 B4JI81 E2A238 E0VML7 B3P448 B4R186 Q9VFU7 B4HFS3 B4PQF3 A0A0N0U2L6 A0A1B6LB71 A0A3L8E1Y0 A0A0L7LS03 A0A195CH45 A0A1B6G259 A0A195AY80 A0A158NPB4 A0A182FA48 A0A232FK68 A0A0L7QTN9 A0A336MDA0 F4WF94 A0A336M8W5
A0A194RK28 A0A194PPB3 A0A2H1WJ89 A0A2A4JRL1 A0A3S2MB94 A0A1B0CF37 A0A0A9ZBH5 A0A0K8T359 A0A1Y1NKS5 D6WPK9 A0A182UG36 A0A1W4WQT7 A0A1W4WRT9 A0A1W4WFY8 A0A1J1HQ06 A0A182LRH8 A0A182KC68 A0A182R5H8 A0A182N5E5 A0A2J7PYK2 A0A194RJS5 A0A067RCL4 A0A1B0BUJ4 N6U7Y1 A0A182X9U7 A0A154P4D6 A0A182L937 A0A182Y570 A0A1Y9IRW5 W5JRL3 A0A182HKS9 A0A182VRA0 A0A182P2F8 A0A182QRL9 A0A182IS29 A0A336MBR2 U4TXN3 A0A2J7PYN3 B0WA44 A0A1B0GLI9 A0A336M9R3 A0A1B6C6V9 A0A0P9ANK1 B4NKE4 A0A1I8PBW5 Q16ZD1 A0A1I8NIL5 A0A0L0BY62 Q7PXI0 A0A0Q9W1D8 A0A1S4GCH1 B4M6A4 A0A0M5JCQ5 A0A1A9UZ31 A0A1W7R8R5 B3LX91 A0A0R3NN42 A0A0A1WVT0 A0A034W9N8 A0A0K8U830 A0A1W4UR97 A0A0K8WMB4 B4G2W0 C0PTV4 W8B9C8 Q297Q6 B4KAA3 A0A1W4V371 A0A3B0KV58 A0A1W4V3H8 A0A1A9Z0I0 B4JI81 E2A238 E0VML7 B3P448 B4R186 Q9VFU7 B4HFS3 B4PQF3 A0A0N0U2L6 A0A1B6LB71 A0A3L8E1Y0 A0A0L7LS03 A0A195CH45 A0A1B6G259 A0A195AY80 A0A158NPB4 A0A182FA48 A0A232FK68 A0A0L7QTN9 A0A336MDA0 F4WF94 A0A336M8W5
Pubmed
26354079
22118469
19121390
25401762
26823975
28004739
+ More
18362917 19820115 24845553 23537049 20966253 25244985 20920257 23761445 17994087 17510324 25315136 26108605 12364791 18057021 15632085 25830018 25348373 24495485 23185243 20798317 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 26227816 21347285 28648823 21719571
18362917 19820115 24845553 23537049 20966253 25244985 20920257 23761445 17994087 17510324 25315136 26108605 12364791 18057021 15632085 25830018 25348373 24495485 23185243 20798317 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 26227816 21347285 28648823 21719571
EMBL
KQ459596
KPI95282.1
AGBW02013965
OWR42507.1
BABH01028139
BABH01028110
+ More
BABH01028111 NWSH01001734 PCG70297.1 AGBW02008416 OWR53449.1 KQ460118 KPJ17690.1 KPI95281.1 ODYU01008707 SOQ52514.1 NWSH01000804 PCG74103.1 RSAL01000001 RVE55293.1 AJWK01009649 AJWK01009650 AJWK01009651 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 GBRD01006221 JAG59600.1 GEZM01002471 JAV97235.1 KQ971354 EFA06826.2 CVRI01000006 CRK88297.1 AXCM01006148 NEVH01020344 PNF21423.1 KPJ17689.1 KK852551 KDR21492.1 JXJN01020724 JXJN01020725 JXJN01020726 APGK01045656 APGK01045657 APGK01045658 APGK01045659 KB741037 ENN74692.1 KQ434809 KZC06717.1 ADMH02000629 ETN65555.1 APCN01000864 AXCN02001176 UFQT01000721 SSX26761.1 KB631716 ERL85567.1 PNF21422.1 DS231868 EDS40777.1 AJWK01035068 AJWK01035069 AJWK01035070 SSX26760.1 GEDC01028050 JAS09248.1 CH902617 KPU79298.1 CH964272 EDW84074.1 CH477492 EAT40016.1 JRES01001160 KNC24967.1 AAAB01008987 EAA01081.5 CH940652 KRF78738.1 EDW59180.1 KRF78739.1 KRF78740.1 CP012526 ALC47181.1 GEHC01000109 JAV47536.1 EDV41691.1 KPU79296.1 KPU79297.1 KPU79299.1 CM000070 KRT00496.1 GBXI01011571 JAD02721.1 GAKP01006691 JAC52261.1 GDHF01029500 JAI22814.1 GDHF01028633 GDHF01000289 JAI23681.1 JAI52025.1 CH479179 EDW24155.1 BT071799 ACN43735.2 GAMC01011313 JAB95242.1 EAL28149.2 KRT00494.1 KRT00495.1 KRT00497.1 CH933806 EDW14590.1 KRG01093.1 KRG01094.1 OUUW01000013 SPP87858.1 CH916369 EDV92962.1 GL435917 EFN72486.1 DS235315 EEB14623.1 CH954181 EDV49222.1 CM000364 EDX13063.1 AE014297 AAF54952.3 AFH06404.1 CH480815 EDW42308.1 CM000160 EDW97249.1 KRK03606.1 KQ436043 KOX67494.1 GEBQ01019020 JAT20957.1 QOIP01000001 RLU26636.1 JTDY01000215 KOB78250.1 KQ977791 KYM99751.1 GECZ01013250 JAS56519.1 KQ976703 KYM77198.1 ADTU01022145 ADTU01022146 ADTU01022147 ADTU01022148 ADTU01022149 ADTU01022150 ADTU01022151 ADTU01022152 NNAY01000092 OXU31062.1 KQ414740 KOC62013.1 SSX26763.1 GL888115 EGI67145.1 SSX26762.1
BABH01028111 NWSH01001734 PCG70297.1 AGBW02008416 OWR53449.1 KQ460118 KPJ17690.1 KPI95281.1 ODYU01008707 SOQ52514.1 NWSH01000804 PCG74103.1 RSAL01000001 RVE55293.1 AJWK01009649 AJWK01009650 AJWK01009651 GBHO01002348 GDHC01008300 JAG41256.1 JAQ10329.1 GBRD01006221 JAG59600.1 GEZM01002471 JAV97235.1 KQ971354 EFA06826.2 CVRI01000006 CRK88297.1 AXCM01006148 NEVH01020344 PNF21423.1 KPJ17689.1 KK852551 KDR21492.1 JXJN01020724 JXJN01020725 JXJN01020726 APGK01045656 APGK01045657 APGK01045658 APGK01045659 KB741037 ENN74692.1 KQ434809 KZC06717.1 ADMH02000629 ETN65555.1 APCN01000864 AXCN02001176 UFQT01000721 SSX26761.1 KB631716 ERL85567.1 PNF21422.1 DS231868 EDS40777.1 AJWK01035068 AJWK01035069 AJWK01035070 SSX26760.1 GEDC01028050 JAS09248.1 CH902617 KPU79298.1 CH964272 EDW84074.1 CH477492 EAT40016.1 JRES01001160 KNC24967.1 AAAB01008987 EAA01081.5 CH940652 KRF78738.1 EDW59180.1 KRF78739.1 KRF78740.1 CP012526 ALC47181.1 GEHC01000109 JAV47536.1 EDV41691.1 KPU79296.1 KPU79297.1 KPU79299.1 CM000070 KRT00496.1 GBXI01011571 JAD02721.1 GAKP01006691 JAC52261.1 GDHF01029500 JAI22814.1 GDHF01028633 GDHF01000289 JAI23681.1 JAI52025.1 CH479179 EDW24155.1 BT071799 ACN43735.2 GAMC01011313 JAB95242.1 EAL28149.2 KRT00494.1 KRT00495.1 KRT00497.1 CH933806 EDW14590.1 KRG01093.1 KRG01094.1 OUUW01000013 SPP87858.1 CH916369 EDV92962.1 GL435917 EFN72486.1 DS235315 EEB14623.1 CH954181 EDV49222.1 CM000364 EDX13063.1 AE014297 AAF54952.3 AFH06404.1 CH480815 EDW42308.1 CM000160 EDW97249.1 KRK03606.1 KQ436043 KOX67494.1 GEBQ01019020 JAT20957.1 QOIP01000001 RLU26636.1 JTDY01000215 KOB78250.1 KQ977791 KYM99751.1 GECZ01013250 JAS56519.1 KQ976703 KYM77198.1 ADTU01022145 ADTU01022146 ADTU01022147 ADTU01022148 ADTU01022149 ADTU01022150 ADTU01022151 ADTU01022152 NNAY01000092 OXU31062.1 KQ414740 KOC62013.1 SSX26763.1 GL888115 EGI67145.1 SSX26762.1
Proteomes
UP000053268
UP000007151
UP000005204
UP000218220
UP000053240
UP000283053
+ More
UP000092461 UP000007266 UP000075902 UP000192223 UP000183832 UP000075883 UP000075881 UP000075900 UP000075884 UP000235965 UP000027135 UP000092460 UP000019118 UP000076407 UP000076502 UP000075882 UP000076408 UP000075903 UP000000673 UP000075840 UP000075920 UP000075885 UP000075886 UP000075880 UP000030742 UP000002320 UP000007801 UP000007798 UP000095300 UP000008820 UP000095301 UP000037069 UP000007062 UP000008792 UP000092553 UP000078200 UP000001819 UP000192221 UP000008744 UP000009192 UP000268350 UP000092445 UP000001070 UP000000311 UP000009046 UP000008711 UP000000304 UP000000803 UP000001292 UP000002282 UP000053105 UP000279307 UP000037510 UP000078542 UP000078540 UP000005205 UP000069272 UP000215335 UP000053825 UP000007755
UP000092461 UP000007266 UP000075902 UP000192223 UP000183832 UP000075883 UP000075881 UP000075900 UP000075884 UP000235965 UP000027135 UP000092460 UP000019118 UP000076407 UP000076502 UP000075882 UP000076408 UP000075903 UP000000673 UP000075840 UP000075920 UP000075885 UP000075886 UP000075880 UP000030742 UP000002320 UP000007801 UP000007798 UP000095300 UP000008820 UP000095301 UP000037069 UP000007062 UP000008792 UP000092553 UP000078200 UP000001819 UP000192221 UP000008744 UP000009192 UP000268350 UP000092445 UP000001070 UP000000311 UP000009046 UP000008711 UP000000304 UP000000803 UP000001292 UP000002282 UP000053105 UP000279307 UP000037510 UP000078542 UP000078540 UP000005205 UP000069272 UP000215335 UP000053825 UP000007755
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194PPP2
A0A212EM07
H9J5G2
H9J5H7
A0A2A4JDY9
A0A212FI98
+ More
A0A194RK28 A0A194PPB3 A0A2H1WJ89 A0A2A4JRL1 A0A3S2MB94 A0A1B0CF37 A0A0A9ZBH5 A0A0K8T359 A0A1Y1NKS5 D6WPK9 A0A182UG36 A0A1W4WQT7 A0A1W4WRT9 A0A1W4WFY8 A0A1J1HQ06 A0A182LRH8 A0A182KC68 A0A182R5H8 A0A182N5E5 A0A2J7PYK2 A0A194RJS5 A0A067RCL4 A0A1B0BUJ4 N6U7Y1 A0A182X9U7 A0A154P4D6 A0A182L937 A0A182Y570 A0A1Y9IRW5 W5JRL3 A0A182HKS9 A0A182VRA0 A0A182P2F8 A0A182QRL9 A0A182IS29 A0A336MBR2 U4TXN3 A0A2J7PYN3 B0WA44 A0A1B0GLI9 A0A336M9R3 A0A1B6C6V9 A0A0P9ANK1 B4NKE4 A0A1I8PBW5 Q16ZD1 A0A1I8NIL5 A0A0L0BY62 Q7PXI0 A0A0Q9W1D8 A0A1S4GCH1 B4M6A4 A0A0M5JCQ5 A0A1A9UZ31 A0A1W7R8R5 B3LX91 A0A0R3NN42 A0A0A1WVT0 A0A034W9N8 A0A0K8U830 A0A1W4UR97 A0A0K8WMB4 B4G2W0 C0PTV4 W8B9C8 Q297Q6 B4KAA3 A0A1W4V371 A0A3B0KV58 A0A1W4V3H8 A0A1A9Z0I0 B4JI81 E2A238 E0VML7 B3P448 B4R186 Q9VFU7 B4HFS3 B4PQF3 A0A0N0U2L6 A0A1B6LB71 A0A3L8E1Y0 A0A0L7LS03 A0A195CH45 A0A1B6G259 A0A195AY80 A0A158NPB4 A0A182FA48 A0A232FK68 A0A0L7QTN9 A0A336MDA0 F4WF94 A0A336M8W5
A0A194RK28 A0A194PPB3 A0A2H1WJ89 A0A2A4JRL1 A0A3S2MB94 A0A1B0CF37 A0A0A9ZBH5 A0A0K8T359 A0A1Y1NKS5 D6WPK9 A0A182UG36 A0A1W4WQT7 A0A1W4WRT9 A0A1W4WFY8 A0A1J1HQ06 A0A182LRH8 A0A182KC68 A0A182R5H8 A0A182N5E5 A0A2J7PYK2 A0A194RJS5 A0A067RCL4 A0A1B0BUJ4 N6U7Y1 A0A182X9U7 A0A154P4D6 A0A182L937 A0A182Y570 A0A1Y9IRW5 W5JRL3 A0A182HKS9 A0A182VRA0 A0A182P2F8 A0A182QRL9 A0A182IS29 A0A336MBR2 U4TXN3 A0A2J7PYN3 B0WA44 A0A1B0GLI9 A0A336M9R3 A0A1B6C6V9 A0A0P9ANK1 B4NKE4 A0A1I8PBW5 Q16ZD1 A0A1I8NIL5 A0A0L0BY62 Q7PXI0 A0A0Q9W1D8 A0A1S4GCH1 B4M6A4 A0A0M5JCQ5 A0A1A9UZ31 A0A1W7R8R5 B3LX91 A0A0R3NN42 A0A0A1WVT0 A0A034W9N8 A0A0K8U830 A0A1W4UR97 A0A0K8WMB4 B4G2W0 C0PTV4 W8B9C8 Q297Q6 B4KAA3 A0A1W4V371 A0A3B0KV58 A0A1W4V3H8 A0A1A9Z0I0 B4JI81 E2A238 E0VML7 B3P448 B4R186 Q9VFU7 B4HFS3 B4PQF3 A0A0N0U2L6 A0A1B6LB71 A0A3L8E1Y0 A0A0L7LS03 A0A195CH45 A0A1B6G259 A0A195AY80 A0A158NPB4 A0A182FA48 A0A232FK68 A0A0L7QTN9 A0A336MDA0 F4WF94 A0A336M8W5
PDB
4OFY
E-value=1.06074e-09,
Score=148
Ontologies
GO
Topology
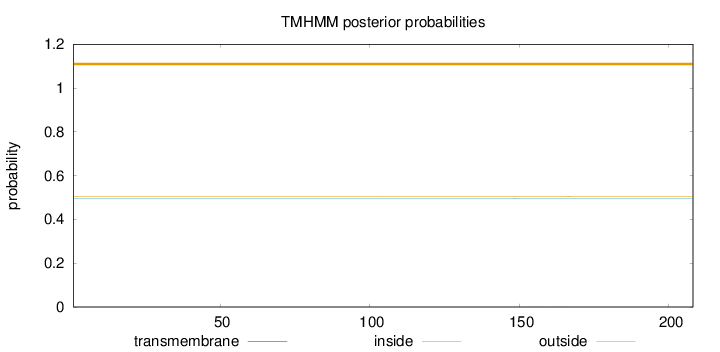
Length:
208
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00566
Exp number, first 60 AAs:
0
Total prob of N-in:
0.49627
outside
1 - 208
Population Genetic Test Statistics
Pi
225.042735
Theta
159.815033
Tajima's D
1.13928
CLR
0.715831
CSRT
0.699715014249288
Interpretation
Uncertain
