Gene
KWMTBOMO15444
Pre Gene Modal
BGIBMGA004770
Annotation
Inwardly_rectifying_k+_channel_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.711
Sequence
CDS
ATGTCAGATTACAACGAAAAATCTGATGAATGGGAAGACGGAAAATTTGTTTTCCAATTCGTTCAAAATTTGGCACCGGGTACATACGTACCTCGTAACAAATTCTATCATCAGATTCCAGTGACAGCAAGAAACAGAAGAGTTTTGTCTAAGGATGGGGAATCTAATTTCTCAAAGCCATTGTTACATTGCCGGAAGGGATTTTCCTACTCTTTCATGACTATAGTCGAAGCCAAATGGAGATGGACCTTATTCTACTTATTTCTGGCCTTCTGCGCAGATTGGTTATTTTTTGGTATAATTTACTGGTTCATCGCACTTGTCCATGGAGACTTATCTGAGGATCATCTGCCTCAATACCAAAACGCAACCGGTTGGGTGCCTTGTATCAAAAATGTCTACGGGTTTACGTCTACATTTCTCTTTAGTATTGAGGTTCACACAACCGTTGCGTACAGTCAAAGGGCGATTACCCCCAACTGCCCAGTAACCATACTAGCTATGTGTCTTCAATGTATAGTCAGTTCTATCTTTCAAGCTTTCATGATCGGGATATTGTTTGCAAAAATAACACGACCCAACGCTAGGGTTGATACTATATTATTCAGCAAACGGGCCGTTATTTCTCTTAGAGACGAGAAATTGTGTTTAATTTTAAGGGTTGGCAATGTGCGGAAATTTAAGATACAAAATGTTAAAGCTACAGTTATTCTATTGAAGTTTATTAATAAGAACAATAACATGTATTTTGTAGAGCAAAAGGAATTAGGTGTTACAATGGATGGATATGAATCGACATTTTCTTTGTGGCCAATATCTGTTGTTCATATTATCGATGAAAAAAGTCCACTTTACGACATATCAGCTGCGGATTTACTATGTGGACATTTCGAAATTTTGGCTATATTCGAAGGTATTAATGGAATCACCGGTCAAGCAGTCCAAGCAAGAACTAGCTACACAGAAAACGATTTGCTTTGGGGTCATCGTTTCGTTCCCATGATAAACGATATAAACAAGATAGCATATGATTTATCTAAACTCAGCGCAACGCATGAAGTAGATACCCCACTGTGCTCGGCTAATGAATATGATGTTTTAATGACGTTAGTTGACTTTGAAGACAAAATATCTGATAAGAAATAG
Protein
MSDYNEKSDEWEDGKFVFQFVQNLAPGTYVPRNKFYHQIPVTARNRRVLSKDGESNFSKPLLHCRKGFSYSFMTIVEAKWRWTLFYLFLAFCADWLFFGIIYWFIALVHGDLSEDHLPQYQNATGWVPCIKNVYGFTSTFLFSIEVHTTVAYSQRAITPNCPVTILAMCLQCIVSSIFQAFMIGILFAKITRPNARVDTILFSKRAVISLRDEKLCLILRVGNVRKFKIQNVKATVILLKFINKNNNMYFVEQKELGVTMDGYESTFSLWPISVVHIIDEKSPLYDISAADLLCGHFEILAIFEGINGITGQAVQARTSYTENDLLWGHRFVPMINDINKIAYDLSKLSATHEVDTPLCSANEYDVLMTLVDFEDKISDKK
Summary
Similarity
Belongs to the inward rectifier-type potassium channel (TC 1.A.2.1) family.
Uniprot
A0A2A4J426
A0A2H1V679
A0A0L7KS10
A0A194RJG2
A0A194PVX3
A0A212EXC8
+ More
H9JC25 A0A1E1W6K4 A0A194PIY2 A0A212FIV9 A0A194RA99 A0A2H1VCZ2 A0A3S2NCQ4 A0A2M4BR23 A0A084W0N0 A0A2A4J5D1 A0A0P6G419 A0A2M4AH79 A0A0P5J2Y0 A0A2M4BN81 A0A2M4BNE8 A0A0L7LKP4 A0A0P5R2X0 A0A182JDE4 A0A2M4BN11 A0A0P5RFU0 A0A0P5UZI0 A0A0P5PAF8 A0A0P5JUR4 A0A0P5LDY8 A0A2M3Z3W1 A0A0N8BGH4 A0A0P6H574 A0A0P4ZEZ9 A0A0P5VXE0 A0A0P5ZZG8 A0A0P5RIK3 A0A0P5USG4 A0A2M3ZH10 A0A0P5WQ73 A0A0P5NSU1 T1EA29 A0A0P5QB03 F5HMU3 A0A1Y9J120 Q16XB6 A0A0P5NEE6 A0A182TWH8 E2A7T2 A0A0P5PAT8 A0A182TA50 A0A0P5LHD3 A0A1Y9IV43 A0A1Y1NIH2 A0A1Y9J0B9 A0A1Y1NJY8 A0A0N8DDX6 A0A182J0B7 A7UVQ1 A0A182MQE6 A0A195FXP7 A0A0P5BAF1 A0A084W0M9 A0A1Y9IVW0 A0A0P6J8U7 A0A0N0U587 A0A0P5D3E7 A0A0P4YDG6 A0A182VGL1 A0A158NMU5 E9FXS0 A0A1Y9J0K5 A0A1C7CZS3 A0A2C9GQR0 Q7PKK8 A0A182M6E0 A0A151IS07 Q16XB7 A0A1Y9J109 E2BHE4 A0A023ES54 A0A182V716 A0A310S9C3 Q7PX91 A0A182UKL8 A0A2M4CNC0 A0A226DFW5 A0A182L8Q7 A0A2C9GQH5 A0A1Y9IVK2 A0A195C1D6 A0A0P5KLK6 A0A1C9TA80 A0A2M4CNA8 A0A1Q3FUZ4 A0A151X0B2 A0A1Y9J115 A0A1Q3FUB8 Q16XB8
H9JC25 A0A1E1W6K4 A0A194PIY2 A0A212FIV9 A0A194RA99 A0A2H1VCZ2 A0A3S2NCQ4 A0A2M4BR23 A0A084W0N0 A0A2A4J5D1 A0A0P6G419 A0A2M4AH79 A0A0P5J2Y0 A0A2M4BN81 A0A2M4BNE8 A0A0L7LKP4 A0A0P5R2X0 A0A182JDE4 A0A2M4BN11 A0A0P5RFU0 A0A0P5UZI0 A0A0P5PAF8 A0A0P5JUR4 A0A0P5LDY8 A0A2M3Z3W1 A0A0N8BGH4 A0A0P6H574 A0A0P4ZEZ9 A0A0P5VXE0 A0A0P5ZZG8 A0A0P5RIK3 A0A0P5USG4 A0A2M3ZH10 A0A0P5WQ73 A0A0P5NSU1 T1EA29 A0A0P5QB03 F5HMU3 A0A1Y9J120 Q16XB6 A0A0P5NEE6 A0A182TWH8 E2A7T2 A0A0P5PAT8 A0A182TA50 A0A0P5LHD3 A0A1Y9IV43 A0A1Y1NIH2 A0A1Y9J0B9 A0A1Y1NJY8 A0A0N8DDX6 A0A182J0B7 A7UVQ1 A0A182MQE6 A0A195FXP7 A0A0P5BAF1 A0A084W0M9 A0A1Y9IVW0 A0A0P6J8U7 A0A0N0U587 A0A0P5D3E7 A0A0P4YDG6 A0A182VGL1 A0A158NMU5 E9FXS0 A0A1Y9J0K5 A0A1C7CZS3 A0A2C9GQR0 Q7PKK8 A0A182M6E0 A0A151IS07 Q16XB7 A0A1Y9J109 E2BHE4 A0A023ES54 A0A182V716 A0A310S9C3 Q7PX91 A0A182UKL8 A0A2M4CNC0 A0A226DFW5 A0A182L8Q7 A0A2C9GQH5 A0A1Y9IVK2 A0A195C1D6 A0A0P5KLK6 A0A1C9TA80 A0A2M4CNA8 A0A1Q3FUZ4 A0A151X0B2 A0A1Y9J115 A0A1Q3FUB8 Q16XB8
Pubmed
EMBL
NWSH01003174
PCG66825.1
ODYU01000868
SOQ36276.1
JTDY01006581
KOB65870.1
+ More
KQ460118 KPJ17692.1 KQ459596 KPI95280.1 AGBW02011785 OWR46149.1 BABH01014775 GDQN01008428 JAT82626.1 KQ459603 KPI93008.1 AGBW02008324 OWR53664.1 KQ460685 KPJ12786.1 ODYU01001885 SOQ38719.1 RSAL01000218 RVE44089.1 GGFJ01006368 MBW55509.1 ATLV01019123 KE525262 KFB43774.1 NWSH01002898 PCG67347.1 GDIQ01038663 JAN56074.1 GGFK01006647 MBW39968.1 GDIQ01211901 JAK39824.1 GGFJ01005331 MBW54472.1 GGFJ01005330 MBW54471.1 JTDY01000713 KOB76123.1 GDIQ01106445 JAL45281.1 GGFJ01005329 MBW54470.1 GDIQ01110646 JAL41080.1 GDIP01109340 JAL94374.1 GDIQ01135575 JAL16151.1 GDIQ01219308 JAK32417.1 GDIQ01171063 JAK80662.1 GGFM01002458 MBW23209.1 GDIQ01180067 JAK71658.1 GDIQ01034371 JAN60366.1 GDIP01226976 JAI96425.1 GDIP01094752 JAM08963.1 GDIP01037108 JAM66607.1 GDIQ01106446 JAL45280.1 GDIP01112339 JAL91375.1 GGFM01007085 MBW27836.1 GDIP01083148 JAM20567.1 GDIQ01159733 JAK91992.1 GAMD01000792 JAB00799.1 GDIQ01123340 JAL28386.1 AAAB01008987 EGK97615.1 KF686745 CH477543 AIN76713.1 EAT39257.1 GDIQ01146529 JAL05197.1 GL437384 EFN70512.1 GDIQ01152930 JAK98795.1 GDIQ01171064 JAK80661.1 GEZM01004174 JAV96445.1 GEZM01004173 JAV96446.1 GDIP01043239 JAM60476.1 EDO63275.2 AXCM01007933 KQ981200 KYN45082.1 GDIP01202629 JAJ20773.1 KFB43773.1 GDIQ01021378 JAN73359.1 KQ435791 KOX74402.1 GDIP01165974 JAJ57428.1 GDIP01229076 JAI94325.1 ADTU01002433 ADTU01002434 ADTU01002435 ADTU01002436 ADTU01002437 GL732526 EFX88146.1 APCN01000829 EAA43222.5 KQ981113 KYN09394.1 KF686744 AIN76712.1 EAT39256.1 GL448287 EFN84913.1 GAPW01001525 JAC12073.1 KQ767621 OAD53388.1 EAA01723.5 GGFL01002636 MBW66814.1 LNIX01000021 OXA43577.1 KQ978350 KYM94689.1 GDIQ01183656 JAK68069.1 KU681439 AOR07219.1 GGFL01002581 MBW66759.1 GFDL01003615 JAV31430.1 KQ982617 KYQ53796.1 GFDL01003851 JAV31194.1 KF686746 AIN76714.1 EAT39258.1
KQ460118 KPJ17692.1 KQ459596 KPI95280.1 AGBW02011785 OWR46149.1 BABH01014775 GDQN01008428 JAT82626.1 KQ459603 KPI93008.1 AGBW02008324 OWR53664.1 KQ460685 KPJ12786.1 ODYU01001885 SOQ38719.1 RSAL01000218 RVE44089.1 GGFJ01006368 MBW55509.1 ATLV01019123 KE525262 KFB43774.1 NWSH01002898 PCG67347.1 GDIQ01038663 JAN56074.1 GGFK01006647 MBW39968.1 GDIQ01211901 JAK39824.1 GGFJ01005331 MBW54472.1 GGFJ01005330 MBW54471.1 JTDY01000713 KOB76123.1 GDIQ01106445 JAL45281.1 GGFJ01005329 MBW54470.1 GDIQ01110646 JAL41080.1 GDIP01109340 JAL94374.1 GDIQ01135575 JAL16151.1 GDIQ01219308 JAK32417.1 GDIQ01171063 JAK80662.1 GGFM01002458 MBW23209.1 GDIQ01180067 JAK71658.1 GDIQ01034371 JAN60366.1 GDIP01226976 JAI96425.1 GDIP01094752 JAM08963.1 GDIP01037108 JAM66607.1 GDIQ01106446 JAL45280.1 GDIP01112339 JAL91375.1 GGFM01007085 MBW27836.1 GDIP01083148 JAM20567.1 GDIQ01159733 JAK91992.1 GAMD01000792 JAB00799.1 GDIQ01123340 JAL28386.1 AAAB01008987 EGK97615.1 KF686745 CH477543 AIN76713.1 EAT39257.1 GDIQ01146529 JAL05197.1 GL437384 EFN70512.1 GDIQ01152930 JAK98795.1 GDIQ01171064 JAK80661.1 GEZM01004174 JAV96445.1 GEZM01004173 JAV96446.1 GDIP01043239 JAM60476.1 EDO63275.2 AXCM01007933 KQ981200 KYN45082.1 GDIP01202629 JAJ20773.1 KFB43773.1 GDIQ01021378 JAN73359.1 KQ435791 KOX74402.1 GDIP01165974 JAJ57428.1 GDIP01229076 JAI94325.1 ADTU01002433 ADTU01002434 ADTU01002435 ADTU01002436 ADTU01002437 GL732526 EFX88146.1 APCN01000829 EAA43222.5 KQ981113 KYN09394.1 KF686744 AIN76712.1 EAT39256.1 GL448287 EFN84913.1 GAPW01001525 JAC12073.1 KQ767621 OAD53388.1 EAA01723.5 GGFL01002636 MBW66814.1 LNIX01000021 OXA43577.1 KQ978350 KYM94689.1 GDIQ01183656 JAK68069.1 KU681439 AOR07219.1 GGFL01002581 MBW66759.1 GFDL01003615 JAV31430.1 KQ982617 KYQ53796.1 GFDL01003851 JAV31194.1 KF686746 AIN76714.1 EAT39258.1
Proteomes
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
UP000005204
+ More
UP000283053 UP000030765 UP000075880 UP000007062 UP000076407 UP000008820 UP000075902 UP000000311 UP000075901 UP000075920 UP000075883 UP000078541 UP000053105 UP000075903 UP000005205 UP000000305 UP000075882 UP000075840 UP000078492 UP000008237 UP000198287 UP000078542 UP000075809
UP000283053 UP000030765 UP000075880 UP000007062 UP000076407 UP000008820 UP000075902 UP000000311 UP000075901 UP000075920 UP000075883 UP000078541 UP000053105 UP000075903 UP000005205 UP000000305 UP000075882 UP000075840 UP000078492 UP000008237 UP000198287 UP000078542 UP000075809
PRIDE
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
A0A2A4J426
A0A2H1V679
A0A0L7KS10
A0A194RJG2
A0A194PVX3
A0A212EXC8
+ More
H9JC25 A0A1E1W6K4 A0A194PIY2 A0A212FIV9 A0A194RA99 A0A2H1VCZ2 A0A3S2NCQ4 A0A2M4BR23 A0A084W0N0 A0A2A4J5D1 A0A0P6G419 A0A2M4AH79 A0A0P5J2Y0 A0A2M4BN81 A0A2M4BNE8 A0A0L7LKP4 A0A0P5R2X0 A0A182JDE4 A0A2M4BN11 A0A0P5RFU0 A0A0P5UZI0 A0A0P5PAF8 A0A0P5JUR4 A0A0P5LDY8 A0A2M3Z3W1 A0A0N8BGH4 A0A0P6H574 A0A0P4ZEZ9 A0A0P5VXE0 A0A0P5ZZG8 A0A0P5RIK3 A0A0P5USG4 A0A2M3ZH10 A0A0P5WQ73 A0A0P5NSU1 T1EA29 A0A0P5QB03 F5HMU3 A0A1Y9J120 Q16XB6 A0A0P5NEE6 A0A182TWH8 E2A7T2 A0A0P5PAT8 A0A182TA50 A0A0P5LHD3 A0A1Y9IV43 A0A1Y1NIH2 A0A1Y9J0B9 A0A1Y1NJY8 A0A0N8DDX6 A0A182J0B7 A7UVQ1 A0A182MQE6 A0A195FXP7 A0A0P5BAF1 A0A084W0M9 A0A1Y9IVW0 A0A0P6J8U7 A0A0N0U587 A0A0P5D3E7 A0A0P4YDG6 A0A182VGL1 A0A158NMU5 E9FXS0 A0A1Y9J0K5 A0A1C7CZS3 A0A2C9GQR0 Q7PKK8 A0A182M6E0 A0A151IS07 Q16XB7 A0A1Y9J109 E2BHE4 A0A023ES54 A0A182V716 A0A310S9C3 Q7PX91 A0A182UKL8 A0A2M4CNC0 A0A226DFW5 A0A182L8Q7 A0A2C9GQH5 A0A1Y9IVK2 A0A195C1D6 A0A0P5KLK6 A0A1C9TA80 A0A2M4CNA8 A0A1Q3FUZ4 A0A151X0B2 A0A1Y9J115 A0A1Q3FUB8 Q16XB8
H9JC25 A0A1E1W6K4 A0A194PIY2 A0A212FIV9 A0A194RA99 A0A2H1VCZ2 A0A3S2NCQ4 A0A2M4BR23 A0A084W0N0 A0A2A4J5D1 A0A0P6G419 A0A2M4AH79 A0A0P5J2Y0 A0A2M4BN81 A0A2M4BNE8 A0A0L7LKP4 A0A0P5R2X0 A0A182JDE4 A0A2M4BN11 A0A0P5RFU0 A0A0P5UZI0 A0A0P5PAF8 A0A0P5JUR4 A0A0P5LDY8 A0A2M3Z3W1 A0A0N8BGH4 A0A0P6H574 A0A0P4ZEZ9 A0A0P5VXE0 A0A0P5ZZG8 A0A0P5RIK3 A0A0P5USG4 A0A2M3ZH10 A0A0P5WQ73 A0A0P5NSU1 T1EA29 A0A0P5QB03 F5HMU3 A0A1Y9J120 Q16XB6 A0A0P5NEE6 A0A182TWH8 E2A7T2 A0A0P5PAT8 A0A182TA50 A0A0P5LHD3 A0A1Y9IV43 A0A1Y1NIH2 A0A1Y9J0B9 A0A1Y1NJY8 A0A0N8DDX6 A0A182J0B7 A7UVQ1 A0A182MQE6 A0A195FXP7 A0A0P5BAF1 A0A084W0M9 A0A1Y9IVW0 A0A0P6J8U7 A0A0N0U587 A0A0P5D3E7 A0A0P4YDG6 A0A182VGL1 A0A158NMU5 E9FXS0 A0A1Y9J0K5 A0A1C7CZS3 A0A2C9GQR0 Q7PKK8 A0A182M6E0 A0A151IS07 Q16XB7 A0A1Y9J109 E2BHE4 A0A023ES54 A0A182V716 A0A310S9C3 Q7PX91 A0A182UKL8 A0A2M4CNC0 A0A226DFW5 A0A182L8Q7 A0A2C9GQH5 A0A1Y9IVK2 A0A195C1D6 A0A0P5KLK6 A0A1C9TA80 A0A2M4CNA8 A0A1Q3FUZ4 A0A151X0B2 A0A1Y9J115 A0A1Q3FUB8 Q16XB8
PDB
3SPI
E-value=5.39739e-61,
Score=594
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
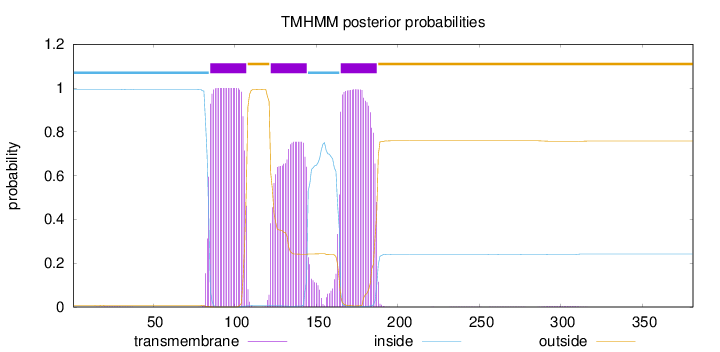
Length:
381
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
62.5421600000001
Exp number, first 60 AAs:
0.02504
Total prob of N-in:
0.99249
inside
1 - 84
TMhelix
85 - 107
outside
108 - 121
TMhelix
122 - 144
inside
145 - 164
TMhelix
165 - 187
outside
188 - 381
Population Genetic Test Statistics
Pi
214.083138
Theta
189.069444
Tajima's D
0.423153
CLR
0.094062
CSRT
0.499075046247688
Interpretation
Uncertain
