Gene
KWMTBOMO15442
Annotation
PREDICTED:_uncharacterized_protein_LOC106713162_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 3.228
Sequence
CDS
ATGAATATTCTTGCCACAAAACACGAACAAAAAGACTTCCGAGGTTTCTGGAAACAAACAAGGAGATTGAGTCCAAAAGCTGGTATCCCGGTGAGCATTGATGGCGAGTGTGAGCCTGTGAGGATTGCTAATCTGTTTAGGAACCATTTCAGGGTACAATCACCCGGTCAGTTAAGTAGAGGTGCTGATGCTGGGGTGGTGGACCCGAGTCGACCCTCGCAAGTACCACCTCGGATAACAACGAAGGAAATAACTAAAATTATAATGAATATGACTAAGGGTAAGTCTCCCGGTCATGACCACCTCAGCATTGAGCACTTTCAACACGCCGGTCCGCATTTACCTAGATTGCTAGCGCTCTTATATACCTTCTGTATCAGACATTCGTATCTGCCCGGGGCGTTAATGAGAACGGTCGTTGTCCCGATTTTAAAAAATAAAACAGGCGACGTATCGGATCGTAACAACTATCGGCCGATCTCCCTGGCAACCATAACTGCTAAGGTACTCGATGCAGTGCTCGAGCGACAGGTGAGTGATCATCTTAAATTAAGTGATGCTCAGTTCGGTTTTAGACCGATGTTATCTACCGAGTGTGCAATCCTGTCGCTAAAGCATGTGGTGGGATACTACACGGAGAAGAATACGCCTGTTTATGCAGTGTTTCTCGATCTTTCGAAGGCTTTCGACCTAGTCAGGTACGACTTGTTGTGGGGTAAGTTGAGGCAGACGGGCGTACCGGGACCTTACGTGGACTTGTTCTCCCATTGGTACGAGTATCAGACCAATCAGGTAAGATGGTCAAACACACTTTCGGAGGAGTACAGGTTGGAATGCGGGGTGAGGCAGGGGGGCTTAACCTCACCGGCGCTTTTCAACCTGTATATTAATGAGTTGATCGAGGCACTCGGTGAGGCACGTGTCGGCTGCTCGATTGATGGTCACTGTGTGAACAGCATCAGTTACGCTGACGACATGGTGCTGTTGAGCCCATCGATCGGTGCTCTCCGGCGGTTGTTGTCCATCTGTGAGAGGTACGCGGAATCTCACGGGCTCCGCTATAATGTTAAAAAAAGTGAACTGCTCGTCTTTAAGGCAGCAAAGACAAAACCGAGTTATGTGCCACCAGTGACGCTCGGAGGTACTCCACTGAAGAGGGTTACTGAGTTCAAATACCTTGGCCATATTGTAACTGAGCACCTTAAAGATGATGCAGATATTGAAAGGGAACGGAGAGCGTTGGCTGTCCGCTGCAGTATGCTGGCCCGCAGGTTCTACCGTTGTACCGCACAGGTTAAGGATGTTGTTGGGGCTCCCGCGGTACTGTAG
Protein
MNILATKHEQKDFRGFWKQTRRLSPKAGIPVSIDGECEPVRIANLFRNHFRVQSPGQLSRGADAGVVDPSRPSQVPPRITTKEITKIIMNMTKGKSPGHDHLSIEHFQHAGPHLPRLLALLYTFCIRHSYLPGALMRTVVVPILKNKTGDVSDRNNYRPISLATITAKVLDAVLERQVSDHLKLSDAQFGFRPMLSTECAILSLKHVVGYYTEKNTPVYAVFLDLSKAFDLVRYDLLWGKLRQTGVPGPYVDLFSHWYEYQTNQVRWSNTLSEEYRLECGVRQGGLTSPALFNLYINELIEALGEARVGCSIDGHCVNSISYADDMVLLSPSIGALRRLLSICERYAESHGLRYNVKKSELLVFKAAKTKPSYVPPVTLGGTPLKRVTEFKYLGHIVTEHLKDDADIERERRALAVRCSMLARRFYRCTAQVKDVVGAPAVL
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
S4NNA8
A0A2A4KA90
A0A2W1BJX4
A0A3S2PB24
A0A2W1BUS0
A0A2W1BGX6
+ More
A0A3S2NHF1 A0A2W1CP14 A0A2W1BE63 A0A2W1BC01 A0A2W1BS65 A0A3S2NQP0 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3B3I0P1 A0A3P9KQQ7 A0A3P8PZH5 A0A1A8UAJ7 A0A146ZPI4 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A3P9K991 A0A0P4VXS2 A0A3C1S1R4 A0A3C1S3F5 A0A3C1S0J5 A0A354GI36 A0A3M7SXU2 A0A336LYA8 W4YPF0 A0A151SQ16 A0A016TUJ4 A0A1Y1K7T4 A0A016UEG0 A0A151QVZ1 A0A016U1Y9 A0A3S0ZV17 A0A3S4QVV5 A0A016VIW8 A0A224XHH0 W4Z133 A0A147BCJ7 A0A3B3H3S6 A0A3B1JC70 A0A2S2QAF2 A0A2H9T4I8 W4YB77 A0A3B3HVG5 A0A3Q3JAD5 D7F178 A0A3C1S4H7 A0A3P9IH04 A0A3B3INA0 A0A016TPK0 A0A3B3IFI4 A0A3B3HSD0 A0A0A9VZM2 A0A146KQ35 A0A0A9XQM2 A0A016WLZ2 A0A3B3HJS0 A0A3P9H8W7 A0A016TPA9 A0A3P9H393 A0A016WMF9 A0A3B3HYZ4 A0A3B3HLK3 A0A3B3IIA9 A0A3B3H5B4 A0A2B7ZXS2 A0A3P9M241 M1BPI1 A0A3B3HTF4 A0A3B3HS69 A0A3P9JDD8 A0A3B3IJY6 A0A3B3H644 A0A3B3HKS7
A0A3S2NHF1 A0A2W1CP14 A0A2W1BE63 A0A2W1BC01 A0A2W1BS65 A0A3S2NQP0 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3B3I0P1 A0A3P9KQQ7 A0A3P8PZH5 A0A1A8UAJ7 A0A146ZPI4 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A3P9K991 A0A0P4VXS2 A0A3C1S1R4 A0A3C1S3F5 A0A3C1S0J5 A0A354GI36 A0A3M7SXU2 A0A336LYA8 W4YPF0 A0A151SQ16 A0A016TUJ4 A0A1Y1K7T4 A0A016UEG0 A0A151QVZ1 A0A016U1Y9 A0A3S0ZV17 A0A3S4QVV5 A0A016VIW8 A0A224XHH0 W4Z133 A0A147BCJ7 A0A3B3H3S6 A0A3B1JC70 A0A2S2QAF2 A0A2H9T4I8 W4YB77 A0A3B3HVG5 A0A3Q3JAD5 D7F178 A0A3C1S4H7 A0A3P9IH04 A0A3B3INA0 A0A016TPK0 A0A3B3IFI4 A0A3B3HSD0 A0A0A9VZM2 A0A146KQ35 A0A0A9XQM2 A0A016WLZ2 A0A3B3HJS0 A0A3P9H8W7 A0A016TPA9 A0A3P9H393 A0A016WMF9 A0A3B3HYZ4 A0A3B3HLK3 A0A3B3IIA9 A0A3B3H5B4 A0A2B7ZXS2 A0A3P9M241 M1BPI1 A0A3B3HTF4 A0A3B3HS69 A0A3P9JDD8 A0A3B3IJY6 A0A3B3H644 A0A3B3HKS7
Pubmed
EMBL
GAIX01014011
JAA78549.1
NWSH01000012
PCG80896.1
KZ150039
PZC74591.1
+ More
RSAL01000132 RVE46368.1 KZ149940 PZC76937.1 KZ150185 PZC72467.1 RSAL01004459 RVE40147.1 NHMN01024349 PZC87407.1 KZ150122 PZC73158.1 KZ150625 PZC70440.1 KZ149956 PZC76465.1 RSAL01000010 RVE53771.1 HAEJ01003895 SBS44352.1 GCES01018090 JAR68233.1 GCES01119762 JAQ66560.1 GCES01122004 JAQ64318.1 GCES01120657 JAQ65665.1 GDRN01099830 JAI58698.1 DMQF01000358 HAO15222.1 DMQF01000507 HAO15811.1 DMQF01000051 HAO14134.1 DNUT01000366 HBI40674.1 REGN01000623 RNA40614.1 UFQT01000284 SSX22780.1 AAGJ04127121 AAGJ04127122 AAGJ04127123 CM003613 KYP56875.1 JARK01001412 EYC06431.1 GEZM01094272 GEZM01094271 JAV55745.1 JARK01001379 EYC13704.1 KQ484600 KYP34423.1 JARK01001398 EYC09120.1 RQTK01000700 RUS75926.1 NCKU01002988 RWS08398.1 JARK01001345 EYC27241.1 GFTR01008526 JAW07900.1 AAGJ04171899 GEGO01006907 JAR88497.1 GGMS01004979 MBY74182.1 NSIT01000254 PJE78133.1 AAGJ04093867 FJ265563 ADI61831.1 DMQF01000521 HAO15860.1 JARK01001422 EYC04705.1 GBHO01042545 GBRD01013465 JAG01059.1 JAG52361.1 GDHC01021277 JAP97351.1 GBHO01021345 JAG22259.1 JARK01000194 EYC40844.1 JARK01001423 EYC04580.1 EYC40845.1 PDHN01000949 PGH37547.1
RSAL01000132 RVE46368.1 KZ149940 PZC76937.1 KZ150185 PZC72467.1 RSAL01004459 RVE40147.1 NHMN01024349 PZC87407.1 KZ150122 PZC73158.1 KZ150625 PZC70440.1 KZ149956 PZC76465.1 RSAL01000010 RVE53771.1 HAEJ01003895 SBS44352.1 GCES01018090 JAR68233.1 GCES01119762 JAQ66560.1 GCES01122004 JAQ64318.1 GCES01120657 JAQ65665.1 GDRN01099830 JAI58698.1 DMQF01000358 HAO15222.1 DMQF01000507 HAO15811.1 DMQF01000051 HAO14134.1 DNUT01000366 HBI40674.1 REGN01000623 RNA40614.1 UFQT01000284 SSX22780.1 AAGJ04127121 AAGJ04127122 AAGJ04127123 CM003613 KYP56875.1 JARK01001412 EYC06431.1 GEZM01094272 GEZM01094271 JAV55745.1 JARK01001379 EYC13704.1 KQ484600 KYP34423.1 JARK01001398 EYC09120.1 RQTK01000700 RUS75926.1 NCKU01002988 RWS08398.1 JARK01001345 EYC27241.1 GFTR01008526 JAW07900.1 AAGJ04171899 GEGO01006907 JAR88497.1 GGMS01004979 MBY74182.1 NSIT01000254 PJE78133.1 AAGJ04093867 FJ265563 ADI61831.1 DMQF01000521 HAO15860.1 JARK01001422 EYC04705.1 GBHO01042545 GBRD01013465 JAG01059.1 JAG52361.1 GDHC01021277 JAP97351.1 GBHO01021345 JAG22259.1 JARK01000194 EYC40844.1 JARK01001423 EYC04580.1 EYC40845.1 PDHN01000949 PGH37547.1
Proteomes
PRIDE
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR024989 MFS_assoc_dom
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR033116 TRYPSIN_SER
IPR036790 Frizzled_dom_sf
IPR020067 Frizzled_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001190 SRCR
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR027124 Swc5/CFDP2
IPR000637 HMGI/Y_DNA-bd_CS
IPR024989 MFS_assoc_dom
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR033116 TRYPSIN_SER
IPR036790 Frizzled_dom_sf
IPR020067 Frizzled_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001190 SRCR
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR027124 Swc5/CFDP2
IPR000637 HMGI/Y_DNA-bd_CS
SUPFAM
Gene 3D
ProteinModelPortal
S4NNA8
A0A2A4KA90
A0A2W1BJX4
A0A3S2PB24
A0A2W1BUS0
A0A2W1BGX6
+ More
A0A3S2NHF1 A0A2W1CP14 A0A2W1BE63 A0A2W1BC01 A0A2W1BS65 A0A3S2NQP0 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3B3I0P1 A0A3P9KQQ7 A0A3P8PZH5 A0A1A8UAJ7 A0A146ZPI4 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A3P9K991 A0A0P4VXS2 A0A3C1S1R4 A0A3C1S3F5 A0A3C1S0J5 A0A354GI36 A0A3M7SXU2 A0A336LYA8 W4YPF0 A0A151SQ16 A0A016TUJ4 A0A1Y1K7T4 A0A016UEG0 A0A151QVZ1 A0A016U1Y9 A0A3S0ZV17 A0A3S4QVV5 A0A016VIW8 A0A224XHH0 W4Z133 A0A147BCJ7 A0A3B3H3S6 A0A3B1JC70 A0A2S2QAF2 A0A2H9T4I8 W4YB77 A0A3B3HVG5 A0A3Q3JAD5 D7F178 A0A3C1S4H7 A0A3P9IH04 A0A3B3INA0 A0A016TPK0 A0A3B3IFI4 A0A3B3HSD0 A0A0A9VZM2 A0A146KQ35 A0A0A9XQM2 A0A016WLZ2 A0A3B3HJS0 A0A3P9H8W7 A0A016TPA9 A0A3P9H393 A0A016WMF9 A0A3B3HYZ4 A0A3B3HLK3 A0A3B3IIA9 A0A3B3H5B4 A0A2B7ZXS2 A0A3P9M241 M1BPI1 A0A3B3HTF4 A0A3B3HS69 A0A3P9JDD8 A0A3B3IJY6 A0A3B3H644 A0A3B3HKS7
A0A3S2NHF1 A0A2W1CP14 A0A2W1BE63 A0A2W1BC01 A0A2W1BS65 A0A3S2NQP0 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3B3I0P1 A0A3P9KQQ7 A0A3P8PZH5 A0A1A8UAJ7 A0A146ZPI4 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A3P9K991 A0A0P4VXS2 A0A3C1S1R4 A0A3C1S3F5 A0A3C1S0J5 A0A354GI36 A0A3M7SXU2 A0A336LYA8 W4YPF0 A0A151SQ16 A0A016TUJ4 A0A1Y1K7T4 A0A016UEG0 A0A151QVZ1 A0A016U1Y9 A0A3S0ZV17 A0A3S4QVV5 A0A016VIW8 A0A224XHH0 W4Z133 A0A147BCJ7 A0A3B3H3S6 A0A3B1JC70 A0A2S2QAF2 A0A2H9T4I8 W4YB77 A0A3B3HVG5 A0A3Q3JAD5 D7F178 A0A3C1S4H7 A0A3P9IH04 A0A3B3INA0 A0A016TPK0 A0A3B3IFI4 A0A3B3HSD0 A0A0A9VZM2 A0A146KQ35 A0A0A9XQM2 A0A016WLZ2 A0A3B3HJS0 A0A3P9H8W7 A0A016TPA9 A0A3P9H393 A0A016WMF9 A0A3B3HYZ4 A0A3B3HLK3 A0A3B3IIA9 A0A3B3H5B4 A0A2B7ZXS2 A0A3P9M241 M1BPI1 A0A3B3HTF4 A0A3B3HS69 A0A3P9JDD8 A0A3B3IJY6 A0A3B3H644 A0A3B3HKS7
PDB
5HHL
E-value=2.83803e-08,
Score=140
Ontologies
PANTHER
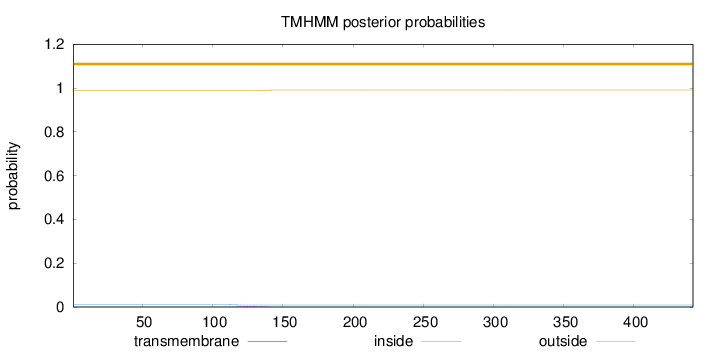
Topology
Length:
442
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0799599999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01238
outside
1 - 442
Population Genetic Test Statistics
Pi
249.472741
Theta
130.579816
Tajima's D
2.960263
CLR
0.384229
CSRT
0.977501124943753
Interpretation
Uncertain
