Gene
KWMTBOMO15439
Pre Gene Modal
BGIBMGA004749
Annotation
PREDICTED:_uncharacterized_protein_LOC101744071_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.787
Sequence
CDS
ATGCGCATTTTACTTATGTTATTACTTACACATTTTGCAACGTCAGCAGTAACTGAAAACCCGTTGCTAATGACAACTGGAAATTCTGATCAGATAGCAAAAACGGCTGAGTGCGTTTTAAAACTATCCGCAAAGTACTTCGTGGAGAGAAAAGCACTAAGCGGCAGCATAGTCATTATCAATGTTAACTCTTATTCGTCTACTACACAAGGGCTACTTTTGAAAACCATACATAGCAGCATAAAATACTCTGTTATGGCGAAGGACTCTTTTTACCCGCATGCGAACGCATCCCATTTCCCGGAGAAAGCCAAAAATTATATGTTGATTCTCGAAGAGAGAACTGAGTTAAAGCGGAACATATTTCAACTGAATAAGTTGCCTAGTTGGAATCCATTAGCTAAAGCTGTAGTTTTTTACCAAATCAAAGGAAACGAGAGTGCGCAACGAATCGCCATCGAGTTTATAAACGAACTGCGAGAACACAAATTTTTCAGGAGTATAATATTCATAAATAATGGAACGGAGTCTGGTGTTACGTCTTATACCTGGCGACCGTACAGTGAAAATAATTGTGGTGGAAAATGCGACTCTGTGTATGTGCTGGATCGGTGTAAAAACAATTTCGTAGAACAGATTGAACCTCAACCCGAATGGTTTCCTTCGAACATGAATGGCTGCCCGTTAACCACATACGCTATTGTTTCTGAACCGTATGTTATGCCCCCAATCAGAAAAATCCCGAATGCCAAATTCGATGACGTTTACGAATTCCAGAAAGGTGGAGAGACTAATTTAGTGAAAACAATCGCAGAGTTCTCCAATATGACATTAATTGTGCGATTATCAGCTATAGAAGAAAATTGGGGTATCATCTATGCGAACGGAACAGCCACCGGGGCTTATGGCGTATTGAGAAATGATTCTGTCGATATAGTTTTTGGGAACATTGAAGTTACCAAACAAATACGAAAATGGTTTCATCCGACAATCAGTTACACTCAAGATGAAATCACTTGGTGTTTACCTAAAGCCGGTCAAGCCTCTGCCTGGGACAACTTAGTCATTATATTCCAATGGACAATCTGGGTGGCGACATTCACTAGTCTCATTTTGATGGGCTTACTTTTTCACTACATGTATTACAGAGAGAAGAATAAGAAAATAACTAAATGGCCAACGAATTCATTACTTATGACTTTCAGTATGCTATTAGGATGGGGTTCGCATTTCGAACCGAAAACCGCCACATTCCGCATTTTGATCTTCGGTTGGCTGTGTTTCAGCATAAACATGGGGATTTCGTATGAGTCGTTCTTAAGAAGTTTCCTAATGCACCCCAGATTCGAGAAACAGATAGCTACAGAGTCAGACCTGATTCAATCTGGAATCCGTTTTGGCGGTAGAGAGATTTATCGTACCTATTTCGAATCTAATGATGCTTCCTCGTCTTATTTGCACACCGAGTATAGCTCCACTACGTTTTCTGAGGGGATAAGGCGTGCCGCTTTGAATCGCGATTTCGCCGTCGTGTCTTCAAGGAGACAGGCTGAGTACCAAGATCAGAAATTAGGGAAAGGTGCGTCGTTGATTTATTGCTTCCCTGAAAGTGATAATTTATATAAATACAGCGTCGTATTGTTGGCTAGGAAATGGTTTCCGATGTTGGAAAGGTTCAACGGTATAATACGTAGCGTTTCCGAGAACGGTTTGATAAATAAATGGAACGATGAGATGTTTATTCATAGGGTTTCGTTGGAGGGAGCGAGTACTATTGTGCCGTTAAGCATTCAGCATTTGTTGGGAGCGTTTATGTTTATTGGTTTCATGTACGGAACTAGTGCTTTTATTTTTCTTGTAGAAGTCTTTGTTGGTTTCGTGCAAAGGAGAGCTTTCTTATCGGCGTTTTTCTGTGGAAAAAAGAAACGTTTTTCAGCTGTTTTTAAAGTTAAAGTGTAG
Protein
MRILLMLLLTHFATSAVTENPLLMTTGNSDQIAKTAECVLKLSAKYFVERKALSGSIVIINVNSYSSTTQGLLLKTIHSSIKYSVMAKDSFYPHANASHFPEKAKNYMLILEERTELKRNIFQLNKLPSWNPLAKAVVFYQIKGNESAQRIAIEFINELREHKFFRSIIFINNGTESGVTSYTWRPYSENNCGGKCDSVYVLDRCKNNFVEQIEPQPEWFPSNMNGCPLTTYAIVSEPYVMPPIRKIPNAKFDDVYEFQKGGETNLVKTIAEFSNMTLIVRLSAIEENWGIIYANGTATGAYGVLRNDSVDIVFGNIEVTKQIRKWFHPTISYTQDEITWCLPKAGQASAWDNLVIIFQWTIWVATFTSLILMGLLFHYMYYREKNKKITKWPTNSLLMTFSMLLGWGSHFEPKTATFRILIFGWLCFSINMGISYESFLRSFLMHPRFEKQIATESDLIQSGIRFGGREIYRTYFESNDASSSYLHTEYSSTTFSEGIRRAALNRDFAVVSSRRQAEYQDQKLGKGASLIYCFPESDNLYKYSVVLLARKWFPMLERFNGIIRSVSENGLINKWNDEMFIHRVSLEGASTIVPLSIQHLLGAFMFIGFMYGTSAFIFLVEVFVGFVQRRAFLSAFFCGKKKRFSAVFKVKV
Summary
Uniprot
H9J5F9
A0A1Y9TJV4
A0A0F7QEJ1
A0A2H1V7G2
A0A3G6V7H8
A0A1V1WCA0
+ More
A0A2A4K3P2 A0A1Q1PP69 A0A0L7KRH6 A0A223HCY3 A0A2K8GL79 H9A5S0 A0A345BF36 A0A3Q8HG95 A0A0K8TUC9 A0A140G9I4 A0A1B3B6Z6 A0A212ETJ1 A0A194PQN6 A0A182MHT5 A0A182RYK2 A0A182K8W1 A0A182VSD2 A0A182QV23 A0A084VLA5 A0A182I7V8 A0A182V2U0 A0A182YGJ7 A0A182XIZ9 A0A182U662 A0A182J2K2 A0A182PF69 A0A182N5P8 A0A1S4FU26 A0A182F6Q5 A0A182GPZ3 A0A182GMM1 Q16PC5 A0A1S4FTY7 B0WEY7 Q16PC4 A0A1J1HH52 A0A1B0CXD1 A0A182GPZ4 A0A1J1HNU9 A0A1J1HII6 A0A336M3L9 A0A182L7G5 A0A182GMM0 A0A1Y1K0Z3 A0A182SAU9 A0A336KKK9 D7EIG1 A0A067QL33 A0A067RMA4 A0A2J7QT11 A0A067RGE0 A0A2P8YNA0 A0A2J7RAK4 A0A2J7PYQ4 A0A2P8XHM3 A0A2P8YQJ2 A0A2J7QH68 A0A2J7PJC3 A0A2P8Y3B4 A0A2P8Z0Q8 A0A067R4T7 A0A2J7R371 A0A0V0J129 A0A2J7Q9U6 A0A1Q1PP76 A0A2P8Z221 A0A2J7RHU9 A0A2P8XKY1 A0A067QXY6 A0A2P8YN99 A0A2P8XZI7 A0A139W8Y9 A0A223HD95 A0A2J7QC88 A0A067R0T1 A0A2J7QQD0 A0A2J7PZG5 A0A2P8Z317 A0A2P8XHL3 A0A2P8XEV5 A0A2P8ZI25 A0A067QYK3 A0A2J7QJK4 A0A2P8XUX9 A0A2P8ZI28 A0A2J7PYG4 A0A067RBR5 A0A2P8ZI31
A0A2A4K3P2 A0A1Q1PP69 A0A0L7KRH6 A0A223HCY3 A0A2K8GL79 H9A5S0 A0A345BF36 A0A3Q8HG95 A0A0K8TUC9 A0A140G9I4 A0A1B3B6Z6 A0A212ETJ1 A0A194PQN6 A0A182MHT5 A0A182RYK2 A0A182K8W1 A0A182VSD2 A0A182QV23 A0A084VLA5 A0A182I7V8 A0A182V2U0 A0A182YGJ7 A0A182XIZ9 A0A182U662 A0A182J2K2 A0A182PF69 A0A182N5P8 A0A1S4FU26 A0A182F6Q5 A0A182GPZ3 A0A182GMM1 Q16PC5 A0A1S4FTY7 B0WEY7 Q16PC4 A0A1J1HH52 A0A1B0CXD1 A0A182GPZ4 A0A1J1HNU9 A0A1J1HII6 A0A336M3L9 A0A182L7G5 A0A182GMM0 A0A1Y1K0Z3 A0A182SAU9 A0A336KKK9 D7EIG1 A0A067QL33 A0A067RMA4 A0A2J7QT11 A0A067RGE0 A0A2P8YNA0 A0A2J7RAK4 A0A2J7PYQ4 A0A2P8XHM3 A0A2P8YQJ2 A0A2J7QH68 A0A2J7PJC3 A0A2P8Y3B4 A0A2P8Z0Q8 A0A067R4T7 A0A2J7R371 A0A0V0J129 A0A2J7Q9U6 A0A1Q1PP76 A0A2P8Z221 A0A2J7RHU9 A0A2P8XKY1 A0A067QXY6 A0A2P8YN99 A0A2P8XZI7 A0A139W8Y9 A0A223HD95 A0A2J7QC88 A0A067R0T1 A0A2J7QQD0 A0A2J7PZG5 A0A2P8Z317 A0A2P8XHL3 A0A2P8XEV5 A0A2P8ZI25 A0A067QYK3 A0A2J7QJK4 A0A2P8XUX9 A0A2P8ZI28 A0A2J7PYG4 A0A067RBR5 A0A2P8ZI31
Pubmed
EMBL
BABH01028180
KX084509
ARO76464.1
LC017796
BAR64810.1
ODYU01001067
+ More
SOQ36790.1 MH193966 AZB49412.1 GENK01000122 JAV45791.1 NWSH01000209 PCG78383.1 KY325458 AQM73619.1 JTDY01006798 KOB65680.1 KY283578 AST36238.1 KY225537 ARO70549.1 JN836720 AFC91760.2 MG820689 AXF48860.1 MF625610 AXY83437.1 GCVX01000143 JAI18087.1 KU702636 AMM70663.1 KT588081 AOE47991.1 AGBW02012567 OWR44808.1 KQ459596 KPI95278.1 AXCM01015069 AXCN02001158 ATLV01014439 KE524972 KFB38749.1 APCN01002560 JXUM01015423 KQ560429 KXJ82457.1 JXUM01001761 KQ560127 KXJ84327.1 CH477788 EAT36213.1 DS231912 EDS25845.1 EAT36214.1 CVRI01000004 CRK87351.1 AJWK01033562 JXUM01015424 KXJ82458.1 CVRI01000006 CRK87905.1 CRK87864.1 UFQT01000509 SSX24852.1 KXJ84326.1 GEZM01097657 JAV54161.1 UFQS01000221 UFQT01000221 SSX01560.1 SSX21940.1 DS497678 EFA11918.1 KK853197 KDR09956.1 KK852427 KDR24123.1 NEVH01011202 PNF31732.1 KK852483 KDR22847.1 PYGN01000473 PSN45722.1 NEVH01006567 PNF37868.1 NEVH01020342 PNF21456.1 PYGN01002101 PSN31492.1 PYGN01000432 PSN46493.1 NEVH01014356 PNF27931.1 NEVH01024949 PNF16419.1 PYGN01000978 PSN38746.1 PSN38753.1 PYGN01000252 PSN50089.1 KK852749 KDR17222.1 NEVH01007822 PNF35288.1 GDKB01000022 JAP38474.1 NEVH01016344 PNF25358.1 KY325453 AQM73614.1 PYGN01000235 PSN50538.1 NEVH01003737 PNF40412.1 PYGN01001824 PSN32653.1 KK853123 KDR11041.1 PSN45721.1 PYGN01001122 PSN37418.1 KQ972879 KXZ75746.1 KY283701 AST36360.1 NEVH01016291 PNF26182.1 KK852798 KDR16369.1 NEVH01012087 PNF30787.1 NEVH01020335 PNF21729.1 PYGN01000219 PSN50894.1 PSN31493.1 PYGN01002421 PSN30525.1 PYGN01000050 PSN56157.1 KK852871 KDR14577.1 NEVH01013553 PNF28758.1 PYGN01001306 PSN35821.1 PSN56158.1 NEVH01020850 PNF21340.1 KK852806 KDR16092.1 PSN56159.1
SOQ36790.1 MH193966 AZB49412.1 GENK01000122 JAV45791.1 NWSH01000209 PCG78383.1 KY325458 AQM73619.1 JTDY01006798 KOB65680.1 KY283578 AST36238.1 KY225537 ARO70549.1 JN836720 AFC91760.2 MG820689 AXF48860.1 MF625610 AXY83437.1 GCVX01000143 JAI18087.1 KU702636 AMM70663.1 KT588081 AOE47991.1 AGBW02012567 OWR44808.1 KQ459596 KPI95278.1 AXCM01015069 AXCN02001158 ATLV01014439 KE524972 KFB38749.1 APCN01002560 JXUM01015423 KQ560429 KXJ82457.1 JXUM01001761 KQ560127 KXJ84327.1 CH477788 EAT36213.1 DS231912 EDS25845.1 EAT36214.1 CVRI01000004 CRK87351.1 AJWK01033562 JXUM01015424 KXJ82458.1 CVRI01000006 CRK87905.1 CRK87864.1 UFQT01000509 SSX24852.1 KXJ84326.1 GEZM01097657 JAV54161.1 UFQS01000221 UFQT01000221 SSX01560.1 SSX21940.1 DS497678 EFA11918.1 KK853197 KDR09956.1 KK852427 KDR24123.1 NEVH01011202 PNF31732.1 KK852483 KDR22847.1 PYGN01000473 PSN45722.1 NEVH01006567 PNF37868.1 NEVH01020342 PNF21456.1 PYGN01002101 PSN31492.1 PYGN01000432 PSN46493.1 NEVH01014356 PNF27931.1 NEVH01024949 PNF16419.1 PYGN01000978 PSN38746.1 PSN38753.1 PYGN01000252 PSN50089.1 KK852749 KDR17222.1 NEVH01007822 PNF35288.1 GDKB01000022 JAP38474.1 NEVH01016344 PNF25358.1 KY325453 AQM73614.1 PYGN01000235 PSN50538.1 NEVH01003737 PNF40412.1 PYGN01001824 PSN32653.1 KK853123 KDR11041.1 PSN45721.1 PYGN01001122 PSN37418.1 KQ972879 KXZ75746.1 KY283701 AST36360.1 NEVH01016291 PNF26182.1 KK852798 KDR16369.1 NEVH01012087 PNF30787.1 NEVH01020335 PNF21729.1 PYGN01000219 PSN50894.1 PSN31493.1 PYGN01002421 PSN30525.1 PYGN01000050 PSN56157.1 KK852871 KDR14577.1 NEVH01013553 PNF28758.1 PYGN01001306 PSN35821.1 PSN56158.1 NEVH01020850 PNF21340.1 KK852806 KDR16092.1 PSN56159.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000075883
+ More
UP000075900 UP000075881 UP000075920 UP000075886 UP000030765 UP000075840 UP000075903 UP000076408 UP000076407 UP000075902 UP000075880 UP000075885 UP000075884 UP000069272 UP000069940 UP000249989 UP000008820 UP000002320 UP000183832 UP000092461 UP000075882 UP000075901 UP000007266 UP000027135 UP000235965 UP000245037
UP000075900 UP000075881 UP000075920 UP000075886 UP000030765 UP000075840 UP000075903 UP000076408 UP000076407 UP000075902 UP000075880 UP000075885 UP000075884 UP000069272 UP000069940 UP000249989 UP000008820 UP000002320 UP000183832 UP000092461 UP000075882 UP000075901 UP000007266 UP000027135 UP000235965 UP000245037
PRIDE
ProteinModelPortal
H9J5F9
A0A1Y9TJV4
A0A0F7QEJ1
A0A2H1V7G2
A0A3G6V7H8
A0A1V1WCA0
+ More
A0A2A4K3P2 A0A1Q1PP69 A0A0L7KRH6 A0A223HCY3 A0A2K8GL79 H9A5S0 A0A345BF36 A0A3Q8HG95 A0A0K8TUC9 A0A140G9I4 A0A1B3B6Z6 A0A212ETJ1 A0A194PQN6 A0A182MHT5 A0A182RYK2 A0A182K8W1 A0A182VSD2 A0A182QV23 A0A084VLA5 A0A182I7V8 A0A182V2U0 A0A182YGJ7 A0A182XIZ9 A0A182U662 A0A182J2K2 A0A182PF69 A0A182N5P8 A0A1S4FU26 A0A182F6Q5 A0A182GPZ3 A0A182GMM1 Q16PC5 A0A1S4FTY7 B0WEY7 Q16PC4 A0A1J1HH52 A0A1B0CXD1 A0A182GPZ4 A0A1J1HNU9 A0A1J1HII6 A0A336M3L9 A0A182L7G5 A0A182GMM0 A0A1Y1K0Z3 A0A182SAU9 A0A336KKK9 D7EIG1 A0A067QL33 A0A067RMA4 A0A2J7QT11 A0A067RGE0 A0A2P8YNA0 A0A2J7RAK4 A0A2J7PYQ4 A0A2P8XHM3 A0A2P8YQJ2 A0A2J7QH68 A0A2J7PJC3 A0A2P8Y3B4 A0A2P8Z0Q8 A0A067R4T7 A0A2J7R371 A0A0V0J129 A0A2J7Q9U6 A0A1Q1PP76 A0A2P8Z221 A0A2J7RHU9 A0A2P8XKY1 A0A067QXY6 A0A2P8YN99 A0A2P8XZI7 A0A139W8Y9 A0A223HD95 A0A2J7QC88 A0A067R0T1 A0A2J7QQD0 A0A2J7PZG5 A0A2P8Z317 A0A2P8XHL3 A0A2P8XEV5 A0A2P8ZI25 A0A067QYK3 A0A2J7QJK4 A0A2P8XUX9 A0A2P8ZI28 A0A2J7PYG4 A0A067RBR5 A0A2P8ZI31
A0A2A4K3P2 A0A1Q1PP69 A0A0L7KRH6 A0A223HCY3 A0A2K8GL79 H9A5S0 A0A345BF36 A0A3Q8HG95 A0A0K8TUC9 A0A140G9I4 A0A1B3B6Z6 A0A212ETJ1 A0A194PQN6 A0A182MHT5 A0A182RYK2 A0A182K8W1 A0A182VSD2 A0A182QV23 A0A084VLA5 A0A182I7V8 A0A182V2U0 A0A182YGJ7 A0A182XIZ9 A0A182U662 A0A182J2K2 A0A182PF69 A0A182N5P8 A0A1S4FU26 A0A182F6Q5 A0A182GPZ3 A0A182GMM1 Q16PC5 A0A1S4FTY7 B0WEY7 Q16PC4 A0A1J1HH52 A0A1B0CXD1 A0A182GPZ4 A0A1J1HNU9 A0A1J1HII6 A0A336M3L9 A0A182L7G5 A0A182GMM0 A0A1Y1K0Z3 A0A182SAU9 A0A336KKK9 D7EIG1 A0A067QL33 A0A067RMA4 A0A2J7QT11 A0A067RGE0 A0A2P8YNA0 A0A2J7RAK4 A0A2J7PYQ4 A0A2P8XHM3 A0A2P8YQJ2 A0A2J7QH68 A0A2J7PJC3 A0A2P8Y3B4 A0A2P8Z0Q8 A0A067R4T7 A0A2J7R371 A0A0V0J129 A0A2J7Q9U6 A0A1Q1PP76 A0A2P8Z221 A0A2J7RHU9 A0A2P8XKY1 A0A067QXY6 A0A2P8YN99 A0A2P8XZI7 A0A139W8Y9 A0A223HD95 A0A2J7QC88 A0A067R0T1 A0A2J7QQD0 A0A2J7PZG5 A0A2P8Z317 A0A2P8XHL3 A0A2P8XEV5 A0A2P8ZI25 A0A067QYK3 A0A2J7QJK4 A0A2P8XUX9 A0A2P8ZI28 A0A2J7PYG4 A0A067RBR5 A0A2P8ZI31
PDB
6CNA
E-value=0.00161859,
Score=100
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.996141
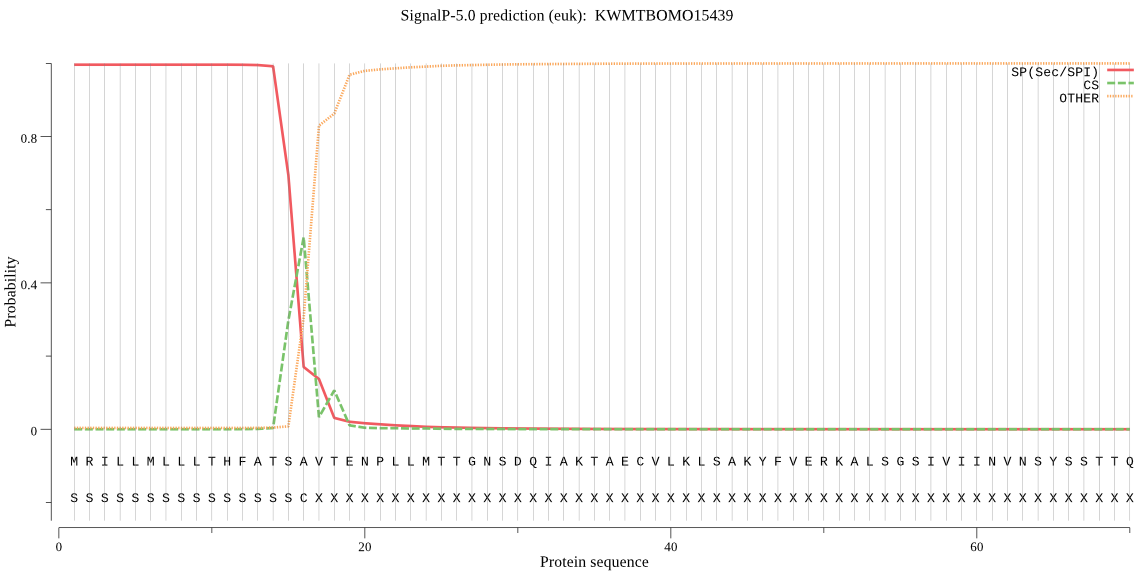
Length:
652
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
76.8241000000001
Exp number, first 60 AAs:
0.03125
Total prob of N-in:
0.00476
outside
1 - 359
TMhelix
360 - 382
inside
383 - 417
TMhelix
418 - 440
outside
441 - 604
TMhelix
605 - 627
inside
628 - 652

Population Genetic Test Statistics
Pi
325.138341
Theta
229.979425
Tajima's D
1.506861
CLR
0
CSRT
0.787960601969901
Interpretation
Uncertain
