Gene
KWMTBOMO15438
Pre Gene Modal
BGIBMGA004748
Annotation
PREDICTED:_neurexin-1-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.114 Nuclear Reliability : 1.521
Sequence
CDS
ATGGGTCTCGGAAACGGTAAGCAGGAGATGCACATCAAACCGTCTAAGACGAGATTCGATGATCATCAGTGGCATAAGTTAACGGTGCACAGAAGGATACAGGAGGTAACTTCGTTCACCAGTTTTTGCAGGGTATCAGCGATAGTCGATGACGTATATTCGGATCATTCCCATGTGGCCGGGTCCTTCACAATGTTGGCGAGTTCAAGGGCTCATGTCGGAGGATCTTTGAATGCCAGAGCCTTGCCCGGTGCCAGGGTCCATACTAATTTTATTGGATGCTTAAAGAAGGTGGAGTTCTCAGCGGATACTCTTCGACTGAACCTCATCGACTTAGCTAGAACGGGCAGCAAATTGATAACCGTAACAGGAAGGTTGGAATACTCCTGCATAGCTACCGACTCTGCGGACCCAGTTACGTTCACCACTAGGGACGCCCATTTGATACTACCAAAATGGGAAGCAGTGAAGACAGGAACAATATCGTTCAAATTTCGAACGAACGAACCGAATGGACTGATCTTATTCAACATGGGCGCCAAGCCACCAAGGGCTGACCTTTTTGCTGTGGAGATCCTCAATGGCTATATCTACGTGCACGTCGATCTTGGATCTGGCGGGGTTAAGGTTCGAGCATCCAGGAGACGGGTGGACGATTCACTCTGGCATGACTTCCTCCTGAGGAGAAGTGGCAGAGACGGCAGAGTTACTGTTGATGGTGCCAACGCTGAGTTTAAGACTCCTGGTGAATCTAATCAGCTGGAGCTGGACGGTCCTCTGTACGTCGGTGGACTCGGTTCAGAGTACTCAGCTTCCAGAACCCCACCGGTTCTTTGGACGGCTGCCCTTAGACAAGGTTTCGTTGGCTGCGTTAGGGACTTGACGCTGAACGGAAAATCTCAGGATCTGACGGCTTTTGCACGACAGCAAGATTCTGCATCAGTGCGCCCAGCCTGCCACGTGCTGATGAAGCAGTGCGTGAGTTCCCCGTGCCAGCACGGCGGGATCTGCTCCGAGGGATGGAACAGACCCCTGTGCAATTGTTCTGCTACGAACTACGGCGGACCGACTTGTGGAAGAGAGTCACCAACAATATTCCTCAACGGCAGCCAGCACCTCACAGCGTCTCTAGGGCCAGAGCACGTGACCCAGACGGAGGAATTAATGCTTCGATTCAGGACCAGTAGGGCTGGTCTCTTGCTGCGAACCGCATCAGAGCATTCGGCGGATCGAATAGAATTGGCCGTAGCTGCTGGTCGGATCAGAGCGAGCGTCAGGCTTGGAGACAGGGAGAAGAACCTTCTAGCAGGAACTAACGTAGCGGACGACAACTGGCACACGGTCCGGTTCTCCCGCCGGGCCTCGAACCTGAGGCTGCAGGTTGACGGGGCGGCCTCGGTGCGCGCCGAAACCATACTAGGCAAGGCGTCGACTTTAGAGATATCGACCCTACATCTCGGCGGATTGTACCACCCGGAGGAGGAAATCCAGATGACCTCGACATTACCAAACTTCGTTGGATACATACAGAAATTCGTTTTCAACGGCATCAAGTATATCGATATGGCCAAAGCACTCGGCGTCGGAAGCGGCGAAGAGCACAACCGTATATACGACACGTCGAATATAATTTTCAACGGGAGATTCGTGAAGCCAGACTCCCTGAACGTTTACAAAGCGGTTACATTCAAATCGAAGCACACGTACGTGGGGCTGCCATTGTTGAAGGCTTACGCGAACACGTATCTAGACTTTTACTTCAGAACGACAGAAATGGACGGTCTGTTGTTTTACAACGGCGGCAAGAAGCAGGATTTCATTGCCATAGAATTAGTGAGCGGGCATGTTCACTGCGTCTTCAATCTGGGCGATGGCGTGGTCACAATGAAGGACAAACTGAAGAACTTCTTGAACGACAACCGATGGCACACCGTGTCGGTCAGACGGCCCACGCCGAAGATACACACACTACAAGTCGACGATGACATCGAAATGCACACGACTAGCTCGAATCTAATGTTGGAACTAGACAGTGTCTTGTACGTAGGAGGCGTGCCCAAGGACATGTATACTGTACTGCCGGTGGGCGTGTTGTCCAGGCAAGGTTTCGAAGGTTGCATGTCCAGCCTTGACCTGCCAGGGGAATCGCCATCATTATTAGAGGATGCCGTTGTACCTAGCTCGTCTTTGGGGTCTGGATGCGAAGGTCCAACGAAGTGCACCCACAACGCATGCGCGAACAAGGGTGTGTGCGTGCAGCAGTGGAACACGTACGTGTGTGATTGCGACCTCACGTCCTTCACTGGGCCCACGTGCTACGACGAATCAATAGCTTACGAATTCGGACCAAGCAGAGGGACAATAACCTACACGTTCCCGCCCAGCTCAGTAGCGGATACGGAGATGGACAAAGTGGCGTTAGGCTTCGTCACCAGCAAGGCAGATGCTGTGTTACTGAAAATCGAGTCTTCAAACACTCAAGATTATATGCAGATGGAGATTATTCGCGGAAACCTTTTCATGGTGTACAACGTTGGTGCTGGTGACCATCCGCTCGGCGACGAGACGACCAGGGTCGATGATGGAGCGTACCACGTGGCGAGGTTTACGAGGAACGGCAGCCGATCGTCGCTCCAGCTCGACAACTATGACGTCATCATTCGGCATCCGCAAGGTGGTCAGCAGTCGACGATCTTCAACAGCATGTCGGTGGTGACGGTGGGCGGCGGCGGCGGCTCGGCGCGCCCGTACAGCGGCGTGCTGGCGGGGCTGGTGGTGGACGGCGTGCGCGTGCTGGAGCTGGCGGCGGCCGGCGACCCCGCCGTGTCCGTGCGCGGTGACGTCAGACGTGCGCACGCGCCGCTCGACAGGGACATCAACAGGATGCAACAGACCCCGCCATCGGGCTACGGAGGGCCGGGAGTACTGGACGAGCTCGTATACTCGGGAGCAGGCTCAGGATGCCGAGATGATGACGAAGACGCGTGCGTACTCCCCGACGCAGGGTCTGGTGATGACCTCATCACTCCCGTTTATGTACCTTCGACTAGGAGACCACCTTCGAAGACGCATAAGGGCGACACGAGTGGTAAGCTGATGAAGCCTTGCGACGACGAGGACTGCATCGAAGGTTCAGGGTCCAACGGCGAAGAGATCACGGAACCAGAACGTCCGGCGACAACGTCGATGTCGTCCAGCACGCACAGCATGACGCCGCCCGCGCCCGCCACCACGGAGCGGCTGCGGCCCGACGTGACGTCACCCGACGTGACGTCACACGAGGTGCCGCGCACCACGCTGCCCGACCACGACAACATCCACGCCGCCACGTTCGGCGTGCACACCGAGACCAACAGAAAGACAACCCCGGACGATCACACGCATACGGCGCATTTCACTCCGACCGATCGCCACGAAGAGGAGCCGACGCACACCACCGCCGAGGACACCAACCACATGCCGGAGTACGAAGACCGCAACGAGAACGAGATAGAAGCTCGCACGCCCACCTACGAGATCGAGGGGGAGCAGGACAGAGCACCCGGGCGAGTCCACCCGGACTACAACGGCTACGAGCACGAGATCACCACCAAGTGGTATCACCCGAAGACGACGGACAACAGAGTCGTGCCGCCAGAGTCGGAGTTCTTTGCCACTATCGTTGGCATAGTGGCGTCGATACTGATAGCCATAATATTGATAGTCATAATTGTGCTCAAATACATCGTGTTCAAGTTCGACCCTTCGTACAAAGTCACAGAGGACAAGAATTACCAGCAAGGAGCCAGCGCCGCGTTGCTAGGCAACCAGAGTCACTCTGGATACCAGACGGGAGTGCAGGGCGGCGTGGCGGCGGGGGGGCAGGGGGGCGCGGGCCGACCCCTGCAGCCGCTCGCCGCGCCCCCTCCCCACCCTGTCAAGAGGGACGGAATCAAGGAGTGGTACGTCTAG
Protein
MGLGNGKQEMHIKPSKTRFDDHQWHKLTVHRRIQEVTSFTSFCRVSAIVDDVYSDHSHVAGSFTMLASSRAHVGGSLNARALPGARVHTNFIGCLKKVEFSADTLRLNLIDLARTGSKLITVTGRLEYSCIATDSADPVTFTTRDAHLILPKWEAVKTGTISFKFRTNEPNGLILFNMGAKPPRADLFAVEILNGYIYVHVDLGSGGVKVRASRRRVDDSLWHDFLLRRSGRDGRVTVDGANAEFKTPGESNQLELDGPLYVGGLGSEYSASRTPPVLWTAALRQGFVGCVRDLTLNGKSQDLTAFARQQDSASVRPACHVLMKQCVSSPCQHGGICSEGWNRPLCNCSATNYGGPTCGRESPTIFLNGSQHLTASLGPEHVTQTEELMLRFRTSRAGLLLRTASEHSADRIELAVAAGRIRASVRLGDREKNLLAGTNVADDNWHTVRFSRRASNLRLQVDGAASVRAETILGKASTLEISTLHLGGLYHPEEEIQMTSTLPNFVGYIQKFVFNGIKYIDMAKALGVGSGEEHNRIYDTSNIIFNGRFVKPDSLNVYKAVTFKSKHTYVGLPLLKAYANTYLDFYFRTTEMDGLLFYNGGKKQDFIAIELVSGHVHCVFNLGDGVVTMKDKLKNFLNDNRWHTVSVRRPTPKIHTLQVDDDIEMHTTSSNLMLELDSVLYVGGVPKDMYTVLPVGVLSRQGFEGCMSSLDLPGESPSLLEDAVVPSSSLGSGCEGPTKCTHNACANKGVCVQQWNTYVCDCDLTSFTGPTCYDESIAYEFGPSRGTITYTFPPSSVADTEMDKVALGFVTSKADAVLLKIESSNTQDYMQMEIIRGNLFMVYNVGAGDHPLGDETTRVDDGAYHVARFTRNGSRSSLQLDNYDVIIRHPQGGQQSTIFNSMSVVTVGGGGGSARPYSGVLAGLVVDGVRVLELAAAGDPAVSVRGDVRRAHAPLDRDINRMQQTPPSGYGGPGVLDELVYSGAGSGCRDDDEDACVLPDAGSGDDLITPVYVPSTRRPPSKTHKGDTSGKLMKPCDDEDCIEGSGSNGEEITEPERPATTSMSSSTHSMTPPAPATTERLRPDVTSPDVTSHEVPRTTLPDHDNIHAATFGVHTETNRKTTPDDHTHTAHFTPTDRHEEEPTHTTAEDTNHMPEYEDRNENEIEARTPTYEIEGEQDRAPGRVHPDYNGYEHEITTKWYHPKTTDNRVVPPESEFFATIVGIVASILIAIILIVIIVLKYIVFKFDPSYKVTEDKNYQQGASAALLGNQSHSGYQTGVQGGVAAGGQGGAGRPLQPLAAPPPHPVKRDGIKEWYV
Summary
Uniprot
H9J5F8
A0A0U2K606
A0A2A4K3Z9
A0A2H1V7J8
A0A194RK32
A0A194PPN7
+ More
A0A212ETK3 A0A1B0CF69 A0A336N0B0 A0A084VES3 Q7PSM9 Q177B3 A0A1I8JU62 A0A1J1J3X1 A0A1I8NI78 A0A2M4CX37 A0A034W6V4 W8B8N0 A0A0L0CEI3 A0A034WAY8 A0A0A1X0M7 A0A0K8VNW6 A0A0K8WLQ9 B4MB86 A0A0K8VCX1 B4NAE7 Q298U9 A0A1W4W394 A0A1Y9J0H9 B4KC19 B3P7H8 Q9VCZ9 B4PMA4 B4HE98 B3MTJ6 A0A3B0K617 A0A3B0K1F4 A0A0Q9X1D3 A0A0R3NKY1 B4JSB3 A0A1W4VQJ7 A0A1W4W1T0 W8AK04 A0A0Q9X2Z6 A5HBQ4 Q3KN41 A0A0R1E473 A0A0R1E4P0 A0A1I8NI73 A0A0Q9WTV7 A0A0R3NH30 A0A3B0K4A6 A0A0P9BMD6 A0A1W4VQK3 A0A0B4KH61 A0A0R1EAE1 A0A3B0KQJ5 A0A0R3NLZ6 A0A0Q9X3A3 A0A0Q9XA33 A0A0Q9X4S7 A0A0B4KGL0 A0A0N8NZ00 A0A0M4EQG8 A0A1W4VQ90 A0A1W4W2N0 A0A0R1E4A5 A0A0P8Y2T0 A0A1W4W1T5 A0A0R1E965 A0A0P8XDQ3 A0A182TUV4 A0A0Q9WSX1 A0A0R3NHJ1 A0A1W4VQ97 A0A0B4KGQ3 A0A0Q9X2W7 A0A3B0K1L2 A0A0R1E499 A0A0P8XEI6 B0X240 A0A336M667 A0A1B0BEH7 A0A182V916 A0A139WFE8 A0A139WFE0 T1HU00 F4WZ00 A0A158NCS4 A0A0C9R3P4 A0A3L8D536 A0A0C9QZT5 K7J1E6 B9VMQ2 A0A0P4Z467
A0A212ETK3 A0A1B0CF69 A0A336N0B0 A0A084VES3 Q7PSM9 Q177B3 A0A1I8JU62 A0A1J1J3X1 A0A1I8NI78 A0A2M4CX37 A0A034W6V4 W8B8N0 A0A0L0CEI3 A0A034WAY8 A0A0A1X0M7 A0A0K8VNW6 A0A0K8WLQ9 B4MB86 A0A0K8VCX1 B4NAE7 Q298U9 A0A1W4W394 A0A1Y9J0H9 B4KC19 B3P7H8 Q9VCZ9 B4PMA4 B4HE98 B3MTJ6 A0A3B0K617 A0A3B0K1F4 A0A0Q9X1D3 A0A0R3NKY1 B4JSB3 A0A1W4VQJ7 A0A1W4W1T0 W8AK04 A0A0Q9X2Z6 A5HBQ4 Q3KN41 A0A0R1E473 A0A0R1E4P0 A0A1I8NI73 A0A0Q9WTV7 A0A0R3NH30 A0A3B0K4A6 A0A0P9BMD6 A0A1W4VQK3 A0A0B4KH61 A0A0R1EAE1 A0A3B0KQJ5 A0A0R3NLZ6 A0A0Q9X3A3 A0A0Q9XA33 A0A0Q9X4S7 A0A0B4KGL0 A0A0N8NZ00 A0A0M4EQG8 A0A1W4VQ90 A0A1W4W2N0 A0A0R1E4A5 A0A0P8Y2T0 A0A1W4W1T5 A0A0R1E965 A0A0P8XDQ3 A0A182TUV4 A0A0Q9WSX1 A0A0R3NHJ1 A0A1W4VQ97 A0A0B4KGQ3 A0A0Q9X2W7 A0A3B0K1L2 A0A0R1E499 A0A0P8XEI6 B0X240 A0A336M667 A0A1B0BEH7 A0A182V916 A0A139WFE8 A0A139WFE0 T1HU00 F4WZ00 A0A158NCS4 A0A0C9R3P4 A0A3L8D536 A0A0C9QZT5 K7J1E6 B9VMQ2 A0A0P4Z467
Pubmed
19121390
26749290
26354079
22118469
24438588
12364791
+ More
14747013 17210077 17510324 25315136 25348373 24495485 26108605 25830018 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17785181 26471740 18362917 19820115 21719571 21347285 30249741 20075255 17069637 18974885
14747013 17210077 17510324 25315136 25348373 24495485 26108605 25830018 17994087 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 17785181 26471740 18362917 19820115 21719571 21347285 30249741 20075255 17069637 18974885
EMBL
BABH01028182
BABH01028183
BABH01028184
KP308210
ALH43428.1
NWSH01000209
+ More
PCG78382.1 ODYU01001067 SOQ36791.1 KQ460118 KPJ17695.1 KQ459596 KPI95277.1 AGBW02012567 OWR44807.1 AJWK01009742 AJWK01009743 AJWK01009744 AJWK01009745 AJWK01009746 AJWK01009747 UFQT01002801 SSX34087.1 ATLV01012289 ATLV01012290 KE524778 KFB36467.1 AAAB01008817 EAA05336.6 CH477379 EAT42236.1 CVRI01000070 CRL07096.1 GGFL01005593 MBW69771.1 GAKP01007666 JAC51286.1 GAMC01020416 GAMC01020415 JAB86139.1 JRES01000501 KNC30666.1 GAKP01007667 JAC51285.1 GBXI01010007 JAD04285.1 GDHF01011733 JAI40581.1 GDHF01000206 JAI52108.1 CH940656 EDW58357.2 GDHF01015612 JAI36702.1 CH964232 EDW80761.1 CM000070 EAL27856.3 CH933806 EDW16892.2 CH954182 EDV54139.1 AE014297 AAF56006.1 AGB96217.1 CM000160 EDW98009.1 CH480815 EDW43195.1 CH902623 EDV30586.1 OUUW01000013 SPP88122.1 SPP88125.1 KRF99331.1 KRT00319.1 CH916373 EDV94653.1 GAMC01020413 JAB86142.1 KRG02364.1 EF460788 ABQ10624.1 BT023898 ABA81832.1 ACZ94983.1 AGB96218.1 AGB96222.1 AHN57466.1 KRK04059.1 KRK04057.1 KRF99332.1 KRT00318.1 SPP88123.1 KPU72939.1 AGB96221.1 KRK04055.1 SPP88126.1 KRT00317.1 KRF99334.1 KRG02362.1 KRG02361.1 AGB96220.1 KPU72940.1 CP012526 ALC47238.1 KRK04056.1 KPU72938.1 KRK04058.1 KPU72936.1 KRF99333.1 KRT00316.1 AGB96219.1 KRG02363.1 SPP88124.1 KRK04054.1 KPU72937.1 DS232279 EDS38970.1 UFQS01000390 UFQT01000390 SSX03502.1 SSX23867.1 JXJN01012903 JXJN01012904 JXJN01012905 JXJN01012906 KQ971352 KYB26674.1 KYB26673.1 ACPB03011671 ACPB03011672 GL888463 EGI60654.1 ADTU01011744 ADTU01011745 ADTU01011746 ADTU01011747 ADTU01011748 ADTU01011749 ADTU01011750 ADTU01011751 ADTU01011752 ADTU01011753 GBYB01007417 JAG77184.1 QOIP01000014 RLU15123.1 GBYB01001230 JAG70997.1 FJ580046 ACM48184.1 GDIP01218212 JAJ05190.1
PCG78382.1 ODYU01001067 SOQ36791.1 KQ460118 KPJ17695.1 KQ459596 KPI95277.1 AGBW02012567 OWR44807.1 AJWK01009742 AJWK01009743 AJWK01009744 AJWK01009745 AJWK01009746 AJWK01009747 UFQT01002801 SSX34087.1 ATLV01012289 ATLV01012290 KE524778 KFB36467.1 AAAB01008817 EAA05336.6 CH477379 EAT42236.1 CVRI01000070 CRL07096.1 GGFL01005593 MBW69771.1 GAKP01007666 JAC51286.1 GAMC01020416 GAMC01020415 JAB86139.1 JRES01000501 KNC30666.1 GAKP01007667 JAC51285.1 GBXI01010007 JAD04285.1 GDHF01011733 JAI40581.1 GDHF01000206 JAI52108.1 CH940656 EDW58357.2 GDHF01015612 JAI36702.1 CH964232 EDW80761.1 CM000070 EAL27856.3 CH933806 EDW16892.2 CH954182 EDV54139.1 AE014297 AAF56006.1 AGB96217.1 CM000160 EDW98009.1 CH480815 EDW43195.1 CH902623 EDV30586.1 OUUW01000013 SPP88122.1 SPP88125.1 KRF99331.1 KRT00319.1 CH916373 EDV94653.1 GAMC01020413 JAB86142.1 KRG02364.1 EF460788 ABQ10624.1 BT023898 ABA81832.1 ACZ94983.1 AGB96218.1 AGB96222.1 AHN57466.1 KRK04059.1 KRK04057.1 KRF99332.1 KRT00318.1 SPP88123.1 KPU72939.1 AGB96221.1 KRK04055.1 SPP88126.1 KRT00317.1 KRF99334.1 KRG02362.1 KRG02361.1 AGB96220.1 KPU72940.1 CP012526 ALC47238.1 KRK04056.1 KPU72938.1 KRK04058.1 KPU72936.1 KRF99333.1 KRT00316.1 AGB96219.1 KRG02363.1 SPP88124.1 KRK04054.1 KPU72937.1 DS232279 EDS38970.1 UFQS01000390 UFQT01000390 SSX03502.1 SSX23867.1 JXJN01012903 JXJN01012904 JXJN01012905 JXJN01012906 KQ971352 KYB26674.1 KYB26673.1 ACPB03011671 ACPB03011672 GL888463 EGI60654.1 ADTU01011744 ADTU01011745 ADTU01011746 ADTU01011747 ADTU01011748 ADTU01011749 ADTU01011750 ADTU01011751 ADTU01011752 ADTU01011753 GBYB01007417 JAG77184.1 QOIP01000014 RLU15123.1 GBYB01001230 JAG70997.1 FJ580046 ACM48184.1 GDIP01218212 JAJ05190.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000092461
+ More
UP000030765 UP000007062 UP000008820 UP000075900 UP000183832 UP000095301 UP000037069 UP000008792 UP000007798 UP000001819 UP000192221 UP000076407 UP000009192 UP000008711 UP000000803 UP000002282 UP000001292 UP000007801 UP000268350 UP000001070 UP000092553 UP000075902 UP000002320 UP000092460 UP000075903 UP000007266 UP000015103 UP000007755 UP000005205 UP000279307 UP000002358
UP000030765 UP000007062 UP000008820 UP000075900 UP000183832 UP000095301 UP000037069 UP000008792 UP000007798 UP000001819 UP000192221 UP000076407 UP000009192 UP000008711 UP000000803 UP000002282 UP000001292 UP000007801 UP000268350 UP000001070 UP000092553 UP000075902 UP000002320 UP000092460 UP000075903 UP000007266 UP000015103 UP000007755 UP000005205 UP000279307 UP000002358
Interpro
SUPFAM
SSF49899
SSF49899
ProteinModelPortal
H9J5F8
A0A0U2K606
A0A2A4K3Z9
A0A2H1V7J8
A0A194RK32
A0A194PPN7
+ More
A0A212ETK3 A0A1B0CF69 A0A336N0B0 A0A084VES3 Q7PSM9 Q177B3 A0A1I8JU62 A0A1J1J3X1 A0A1I8NI78 A0A2M4CX37 A0A034W6V4 W8B8N0 A0A0L0CEI3 A0A034WAY8 A0A0A1X0M7 A0A0K8VNW6 A0A0K8WLQ9 B4MB86 A0A0K8VCX1 B4NAE7 Q298U9 A0A1W4W394 A0A1Y9J0H9 B4KC19 B3P7H8 Q9VCZ9 B4PMA4 B4HE98 B3MTJ6 A0A3B0K617 A0A3B0K1F4 A0A0Q9X1D3 A0A0R3NKY1 B4JSB3 A0A1W4VQJ7 A0A1W4W1T0 W8AK04 A0A0Q9X2Z6 A5HBQ4 Q3KN41 A0A0R1E473 A0A0R1E4P0 A0A1I8NI73 A0A0Q9WTV7 A0A0R3NH30 A0A3B0K4A6 A0A0P9BMD6 A0A1W4VQK3 A0A0B4KH61 A0A0R1EAE1 A0A3B0KQJ5 A0A0R3NLZ6 A0A0Q9X3A3 A0A0Q9XA33 A0A0Q9X4S7 A0A0B4KGL0 A0A0N8NZ00 A0A0M4EQG8 A0A1W4VQ90 A0A1W4W2N0 A0A0R1E4A5 A0A0P8Y2T0 A0A1W4W1T5 A0A0R1E965 A0A0P8XDQ3 A0A182TUV4 A0A0Q9WSX1 A0A0R3NHJ1 A0A1W4VQ97 A0A0B4KGQ3 A0A0Q9X2W7 A0A3B0K1L2 A0A0R1E499 A0A0P8XEI6 B0X240 A0A336M667 A0A1B0BEH7 A0A182V916 A0A139WFE8 A0A139WFE0 T1HU00 F4WZ00 A0A158NCS4 A0A0C9R3P4 A0A3L8D536 A0A0C9QZT5 K7J1E6 B9VMQ2 A0A0P4Z467
A0A212ETK3 A0A1B0CF69 A0A336N0B0 A0A084VES3 Q7PSM9 Q177B3 A0A1I8JU62 A0A1J1J3X1 A0A1I8NI78 A0A2M4CX37 A0A034W6V4 W8B8N0 A0A0L0CEI3 A0A034WAY8 A0A0A1X0M7 A0A0K8VNW6 A0A0K8WLQ9 B4MB86 A0A0K8VCX1 B4NAE7 Q298U9 A0A1W4W394 A0A1Y9J0H9 B4KC19 B3P7H8 Q9VCZ9 B4PMA4 B4HE98 B3MTJ6 A0A3B0K617 A0A3B0K1F4 A0A0Q9X1D3 A0A0R3NKY1 B4JSB3 A0A1W4VQJ7 A0A1W4W1T0 W8AK04 A0A0Q9X2Z6 A5HBQ4 Q3KN41 A0A0R1E473 A0A0R1E4P0 A0A1I8NI73 A0A0Q9WTV7 A0A0R3NH30 A0A3B0K4A6 A0A0P9BMD6 A0A1W4VQK3 A0A0B4KH61 A0A0R1EAE1 A0A3B0KQJ5 A0A0R3NLZ6 A0A0Q9X3A3 A0A0Q9XA33 A0A0Q9X4S7 A0A0B4KGL0 A0A0N8NZ00 A0A0M4EQG8 A0A1W4VQ90 A0A1W4W2N0 A0A0R1E4A5 A0A0P8Y2T0 A0A1W4W1T5 A0A0R1E965 A0A0P8XDQ3 A0A182TUV4 A0A0Q9WSX1 A0A0R3NHJ1 A0A1W4VQ97 A0A0B4KGQ3 A0A0Q9X2W7 A0A3B0K1L2 A0A0R1E499 A0A0P8XEI6 B0X240 A0A336M667 A0A1B0BEH7 A0A182V916 A0A139WFE8 A0A139WFE0 T1HU00 F4WZ00 A0A158NCS4 A0A0C9R3P4 A0A3L8D536 A0A0C9QZT5 K7J1E6 B9VMQ2 A0A0P4Z467
PDB
3QCW
E-value=0,
Score=1869
Ontologies
GO
GO:0016021
GO:0051124
GO:1900073
GO:0007268
GO:1900244
GO:0072553
GO:0019901
GO:0061174
GO:2000331
GO:0040012
GO:0050808
GO:0007274
GO:0048488
GO:0040011
GO:0097109
GO:0007416
GO:0008306
GO:0071938
GO:0031594
GO:0034185
GO:0097105
GO:0097118
GO:0042043
GO:0043195
GO:0099056
GO:0098820
GO:0046872
GO:0005509
GO:0005515
GO:0043169
GO:0000287
GO:0004721
GO:0030145
GO:0043565
GO:0003677
GO:0003707
GO:0006281
GO:0006259
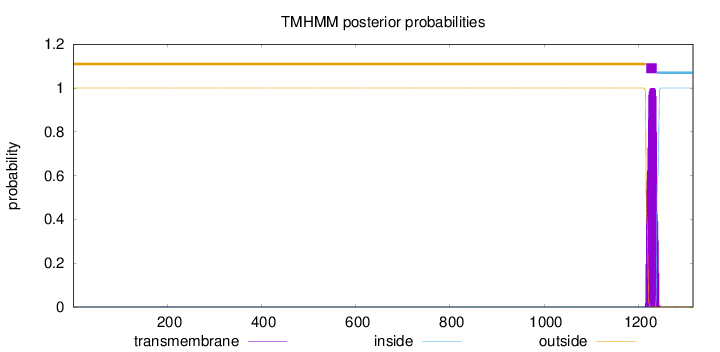
Topology
Length:
1316
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.9728
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.00018
outside
1 - 1216
TMhelix
1217 - 1239
inside
1240 - 1316
Population Genetic Test Statistics
Pi
271.252371
Theta
176.394384
Tajima's D
1.598049
CLR
0.285573
CSRT
0.813409329533523
Interpretation
Uncertain
