Gene
KWMTBOMO15431
Pre Gene Modal
BGIBMGA004745
Annotation
PREDICTED:_protein_preli-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.594
Sequence
CDS
ATGACGAGATATTTCGAAAATACTTCGGTCTTTAACTTCAGTTGGGATCAAGTTGCGCGTGGTTACTGGAGGAGATACCCAAATCCGCAGAGTACTCACGTCCTATCTGAAGATACATGGAGCCGACAAGTTCGCGACGGTTGTCTTTATACAAAGCGGCTCCTGACTAAAACCAACAGAGTTCCAAAATGGGGTGAAAGATTTTTCAACTCTAAATCCGTTAAAATAATCGAGGAGAGTGTCGTTGATCCGGAAAAGAAGATTCTCGTAACATACACCAGAAATTTAGGGTACACTAAAGTGATGAGTGTAGTTGAGAGAGTAGAGTACAGGTCAGCCGGTCCGGGCCAGACCGTGGCCAAGCGCTCGGCGTGGATCGACTCGCAGGTTTTCGGCTTCTCGCGAGCCATACGGGCATTCGGTATCGAAAGGTTCAAGAAGAACTGCTCGCAGATGGCGAGCGGGTTCAACCACGTCCTGCACAGCATGTACCCGCGGCCCGCGAGCCCCGCCTCCGGCATGGCCCTGTCTCTGAAGGAGATGCGCGAGGCGGCCGCCGCGGCCACGGACCGCGCCAAGGCCAACGTGCTCTCGTCCGTGCAGAGGACGTAG
Protein
MTRYFENTSVFNFSWDQVARGYWRRYPNPQSTHVLSEDTWSRQVRDGCLYTKRLLTKTNRVPKWGERFFNSKSVKIIEESVVDPEKKILVTYTRNLGYTKVMSVVERVEYRSAGPGQTVAKRSAWIDSQVFGFSRAIRAFGIERFKKNCSQMASGFNHVLHSMYPRPASPASGMALSLKEMREAAAAATDRAKANVLSSVQRT
Summary
Uniprot
H9J5F5
A0A3S2P2A7
A0A1E1WLS3
A0A212FC11
A0A232F898
K7JPJ5
+ More
A0A1B0C9D2 A0A0K8TSW3 A0A2P8XZ43 A0A1B0DC51 A0A2J7QIP8 A0A067RNI5 A0A084WQZ4 A0A182H1Z1 T1DNV5 F4X6H1 A0A195E6W3 A0A310SA73 E2AM01 W5JWL5 Q1HQR7 A0A158NFZ7 Q16ME3 A0A026W6M9 A0A023F9P8 A0A161M4X0 A0A182INH1 D6WC51 A0A151IQD6 A0A1Q3G468 A0A2M4ASA2 A0A2M4BZB6 A0A182FKE5 A0A182KKD9 Q5TVQ1 A0A182HVU9 A0A224XXE8 A0A336M5R9 E2B3L0 V9IBB4 A0A069DXC2 A0A182WUM6 A0A1Q3G428 A0A2M3ZJI1 A0A1Q3G431 A0A182MB79 A0A182VDL3 A0A182U3S9 A0A0V0G5E1 A0A2A3EED4 V9ID41 A0A0B4J2N8 A0A182YKP1 A0A182PRN0 A0A1L8DVL3 A0A2J7RL08 A0A151XBY2 A0A182QWZ0 A0A195B2T2 A0A182WFV1 A0A1L8DVJ2 E9J609 U5EJY9 A0A0L7QKJ4 A0A0P4VRX9 R4G889 A0A2R7WBD9 A0A1W4XF77 A0A182R2X4 A0A182K195 A0A182NA36 E0V908 A0A0T6BD64 A0A1Y1MZ66 A0A1J1J188 A0A154PHX7 N6TTW3 W8BQJ2 A0A194PQN0 A0A1B6E3G8 A0A0L0BPG1 A0A0A1WV88 A0A0A1X6Q7 A0A0A1XSK0 A0A1D2N8Z7 A0A2H8TPE2 A0A2S2NWB1 A0A034WHY5 A0A1B6FJ75 A0A1I8MPK0 C4WTG8 A0A0K8VYX5 A0A1B6K362 A0A1I8PFW9 A0A1B6LL22 A0A0P4VZF1 A0A1A9WIH7 A0A1B0A1Z4
A0A1B0C9D2 A0A0K8TSW3 A0A2P8XZ43 A0A1B0DC51 A0A2J7QIP8 A0A067RNI5 A0A084WQZ4 A0A182H1Z1 T1DNV5 F4X6H1 A0A195E6W3 A0A310SA73 E2AM01 W5JWL5 Q1HQR7 A0A158NFZ7 Q16ME3 A0A026W6M9 A0A023F9P8 A0A161M4X0 A0A182INH1 D6WC51 A0A151IQD6 A0A1Q3G468 A0A2M4ASA2 A0A2M4BZB6 A0A182FKE5 A0A182KKD9 Q5TVQ1 A0A182HVU9 A0A224XXE8 A0A336M5R9 E2B3L0 V9IBB4 A0A069DXC2 A0A182WUM6 A0A1Q3G428 A0A2M3ZJI1 A0A1Q3G431 A0A182MB79 A0A182VDL3 A0A182U3S9 A0A0V0G5E1 A0A2A3EED4 V9ID41 A0A0B4J2N8 A0A182YKP1 A0A182PRN0 A0A1L8DVL3 A0A2J7RL08 A0A151XBY2 A0A182QWZ0 A0A195B2T2 A0A182WFV1 A0A1L8DVJ2 E9J609 U5EJY9 A0A0L7QKJ4 A0A0P4VRX9 R4G889 A0A2R7WBD9 A0A1W4XF77 A0A182R2X4 A0A182K195 A0A182NA36 E0V908 A0A0T6BD64 A0A1Y1MZ66 A0A1J1J188 A0A154PHX7 N6TTW3 W8BQJ2 A0A194PQN0 A0A1B6E3G8 A0A0L0BPG1 A0A0A1WV88 A0A0A1X6Q7 A0A0A1XSK0 A0A1D2N8Z7 A0A2H8TPE2 A0A2S2NWB1 A0A034WHY5 A0A1B6FJ75 A0A1I8MPK0 C4WTG8 A0A0K8VYX5 A0A1B6K362 A0A1I8PFW9 A0A1B6LL22 A0A0P4VZF1 A0A1A9WIH7 A0A1B0A1Z4
Pubmed
19121390
22118469
28648823
20075255
26369729
29403074
+ More
24845553 24438588 26483478 21719571 20798317 20920257 23761445 17204158 21347285 17510324 24508170 30249741 25474469 18362917 19820115 20966253 12364791 14747013 17210077 26334808 25244985 21282665 27129103 20566863 28004739 23537049 24495485 26354079 26108605 25830018 27289101 25348373 25315136
24845553 24438588 26483478 21719571 20798317 20920257 23761445 17204158 21347285 17510324 24508170 30249741 25474469 18362917 19820115 20966253 12364791 14747013 17210077 26334808 25244985 21282665 27129103 20566863 28004739 23537049 24495485 26354079 26108605 25830018 27289101 25348373 25315136
EMBL
BABH01028201
RSAL01000001
RVE55303.1
GDQN01003129
GDQN01002345
JAT87925.1
+ More
JAT88709.1 AGBW02009250 OWR51263.1 NNAY01000785 OXU26527.1 AAZX01000464 AJWK01002228 GDAI01000382 JAI17221.1 PYGN01001145 PSN37194.1 AJVK01005024 NEVH01013579 PNF28461.1 KK852563 KDR21294.1 ATLV01025855 ATLV01025856 KE525401 KFB52638.1 JXUM01023411 JXUM01023412 JXUM01023413 KQ560661 KXJ81385.1 GAMD01002535 JAA99055.1 GL888818 EGI57931.1 KQ979568 KYN20923.1 KQ780658 OAD51931.1 GL440703 EFN65521.1 ADMH02000135 ETN67645.1 DQ440377 ABF18410.1 ADTU01014868 CH477863 EAT35516.1 KK107373 QOIP01000004 EZA51623.1 RLU23321.1 GBBI01001064 JAC17648.1 GEMB01002659 JAS00533.1 KQ971316 EEZ98761.1 KQ976780 KYN08369.1 GFDL01000486 JAV34559.1 GGFK01010291 MBW43612.1 GGFJ01009242 MBW58383.1 AAAB01008823 EAL41461.2 APCN01001422 GFTR01003583 JAW12843.1 UFQT01000376 SSX23707.1 GL445346 EFN89762.1 JR038745 AEY58390.1 GBGD01002895 JAC85994.1 GFDL01000487 JAV34558.1 GGFM01007930 MBW28681.1 GFDL01000488 JAV34557.1 AXCM01000462 GECL01002844 JAP03280.1 KZ288287 PBC29371.1 JR038746 AEY58391.1 GFDF01003631 JAV10453.1 NEVH01002701 PNF41527.1 KQ982314 KYQ57872.1 AXCN02002378 KQ976662 KYM78590.1 GFDF01003632 JAV10452.1 GL768221 EFZ11712.1 GANO01002021 JAB57850.1 KQ414940 KOC59149.1 GDKW01000768 JAI55827.1 ACPB03013965 GAHY01001392 JAA76118.1 KK854406 PTY15555.1 DS234988 EEB09864.1 LJIG01001664 KRT85276.1 GEZM01017050 GEZM01017049 GEZM01017046 JAV90939.1 CVRI01000065 CRL05698.1 KQ434902 KZC11094.1 APGK01052724 APGK01052725 KB741213 KB631753 ENN72685.1 ERL85808.1 GAMC01011034 GAMC01011033 JAB95521.1 KQ459596 KPI95273.1 GEDC01004834 JAS32464.1 JRES01001654 KNC21119.1 GBXI01011530 JAD02762.1 GBXI01007969 GBXI01002822 GBXI01000412 JAD06323.1 JAD11470.1 JAD13880.1 GBXI01000452 JAD13840.1 LJIJ01000151 ODN01456.1 GFXV01004238 MBW16043.1 GGMR01008723 MBY21342.1 GAKP01004995 GAKP01004994 JAC53957.1 GECZ01019542 JAS50227.1 ABLF02029448 AK340608 BAH71188.1 GDHF01013779 GDHF01008196 GDHF01007063 JAI38535.1 JAI44118.1 JAI45251.1 GECU01016363 GECU01001852 JAS91343.1 JAT05855.1 GEBQ01026854 GEBQ01015638 JAT13123.1 JAT24339.1 GDRN01084801 JAI61438.1
JAT88709.1 AGBW02009250 OWR51263.1 NNAY01000785 OXU26527.1 AAZX01000464 AJWK01002228 GDAI01000382 JAI17221.1 PYGN01001145 PSN37194.1 AJVK01005024 NEVH01013579 PNF28461.1 KK852563 KDR21294.1 ATLV01025855 ATLV01025856 KE525401 KFB52638.1 JXUM01023411 JXUM01023412 JXUM01023413 KQ560661 KXJ81385.1 GAMD01002535 JAA99055.1 GL888818 EGI57931.1 KQ979568 KYN20923.1 KQ780658 OAD51931.1 GL440703 EFN65521.1 ADMH02000135 ETN67645.1 DQ440377 ABF18410.1 ADTU01014868 CH477863 EAT35516.1 KK107373 QOIP01000004 EZA51623.1 RLU23321.1 GBBI01001064 JAC17648.1 GEMB01002659 JAS00533.1 KQ971316 EEZ98761.1 KQ976780 KYN08369.1 GFDL01000486 JAV34559.1 GGFK01010291 MBW43612.1 GGFJ01009242 MBW58383.1 AAAB01008823 EAL41461.2 APCN01001422 GFTR01003583 JAW12843.1 UFQT01000376 SSX23707.1 GL445346 EFN89762.1 JR038745 AEY58390.1 GBGD01002895 JAC85994.1 GFDL01000487 JAV34558.1 GGFM01007930 MBW28681.1 GFDL01000488 JAV34557.1 AXCM01000462 GECL01002844 JAP03280.1 KZ288287 PBC29371.1 JR038746 AEY58391.1 GFDF01003631 JAV10453.1 NEVH01002701 PNF41527.1 KQ982314 KYQ57872.1 AXCN02002378 KQ976662 KYM78590.1 GFDF01003632 JAV10452.1 GL768221 EFZ11712.1 GANO01002021 JAB57850.1 KQ414940 KOC59149.1 GDKW01000768 JAI55827.1 ACPB03013965 GAHY01001392 JAA76118.1 KK854406 PTY15555.1 DS234988 EEB09864.1 LJIG01001664 KRT85276.1 GEZM01017050 GEZM01017049 GEZM01017046 JAV90939.1 CVRI01000065 CRL05698.1 KQ434902 KZC11094.1 APGK01052724 APGK01052725 KB741213 KB631753 ENN72685.1 ERL85808.1 GAMC01011034 GAMC01011033 JAB95521.1 KQ459596 KPI95273.1 GEDC01004834 JAS32464.1 JRES01001654 KNC21119.1 GBXI01011530 JAD02762.1 GBXI01007969 GBXI01002822 GBXI01000412 JAD06323.1 JAD11470.1 JAD13880.1 GBXI01000452 JAD13840.1 LJIJ01000151 ODN01456.1 GFXV01004238 MBW16043.1 GGMR01008723 MBY21342.1 GAKP01004995 GAKP01004994 JAC53957.1 GECZ01019542 JAS50227.1 ABLF02029448 AK340608 BAH71188.1 GDHF01013779 GDHF01008196 GDHF01007063 JAI38535.1 JAI44118.1 JAI45251.1 GECU01016363 GECU01001852 JAS91343.1 JAT05855.1 GEBQ01026854 GEBQ01015638 JAT13123.1 JAT24339.1 GDRN01084801 JAI61438.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000215335
UP000002358
UP000092461
+ More
UP000245037 UP000092462 UP000235965 UP000027135 UP000030765 UP000069940 UP000249989 UP000007755 UP000078492 UP000000311 UP000000673 UP000005205 UP000008820 UP000053097 UP000279307 UP000075880 UP000007266 UP000078542 UP000069272 UP000075882 UP000007062 UP000075840 UP000008237 UP000076407 UP000075883 UP000075903 UP000075902 UP000242457 UP000005203 UP000076408 UP000075885 UP000075809 UP000075886 UP000078540 UP000075920 UP000053825 UP000015103 UP000192223 UP000075900 UP000075881 UP000075884 UP000009046 UP000183832 UP000076502 UP000019118 UP000030742 UP000053268 UP000037069 UP000094527 UP000095301 UP000007819 UP000095300 UP000091820 UP000092445
UP000245037 UP000092462 UP000235965 UP000027135 UP000030765 UP000069940 UP000249989 UP000007755 UP000078492 UP000000311 UP000000673 UP000005205 UP000008820 UP000053097 UP000279307 UP000075880 UP000007266 UP000078542 UP000069272 UP000075882 UP000007062 UP000075840 UP000008237 UP000076407 UP000075883 UP000075903 UP000075902 UP000242457 UP000005203 UP000076408 UP000075885 UP000075809 UP000075886 UP000078540 UP000075920 UP000053825 UP000015103 UP000192223 UP000075900 UP000075881 UP000075884 UP000009046 UP000183832 UP000076502 UP000019118 UP000030742 UP000053268 UP000037069 UP000094527 UP000095301 UP000007819 UP000095300 UP000091820 UP000092445
PRIDE
Pfam
PF04707 PRELI
ProteinModelPortal
H9J5F5
A0A3S2P2A7
A0A1E1WLS3
A0A212FC11
A0A232F898
K7JPJ5
+ More
A0A1B0C9D2 A0A0K8TSW3 A0A2P8XZ43 A0A1B0DC51 A0A2J7QIP8 A0A067RNI5 A0A084WQZ4 A0A182H1Z1 T1DNV5 F4X6H1 A0A195E6W3 A0A310SA73 E2AM01 W5JWL5 Q1HQR7 A0A158NFZ7 Q16ME3 A0A026W6M9 A0A023F9P8 A0A161M4X0 A0A182INH1 D6WC51 A0A151IQD6 A0A1Q3G468 A0A2M4ASA2 A0A2M4BZB6 A0A182FKE5 A0A182KKD9 Q5TVQ1 A0A182HVU9 A0A224XXE8 A0A336M5R9 E2B3L0 V9IBB4 A0A069DXC2 A0A182WUM6 A0A1Q3G428 A0A2M3ZJI1 A0A1Q3G431 A0A182MB79 A0A182VDL3 A0A182U3S9 A0A0V0G5E1 A0A2A3EED4 V9ID41 A0A0B4J2N8 A0A182YKP1 A0A182PRN0 A0A1L8DVL3 A0A2J7RL08 A0A151XBY2 A0A182QWZ0 A0A195B2T2 A0A182WFV1 A0A1L8DVJ2 E9J609 U5EJY9 A0A0L7QKJ4 A0A0P4VRX9 R4G889 A0A2R7WBD9 A0A1W4XF77 A0A182R2X4 A0A182K195 A0A182NA36 E0V908 A0A0T6BD64 A0A1Y1MZ66 A0A1J1J188 A0A154PHX7 N6TTW3 W8BQJ2 A0A194PQN0 A0A1B6E3G8 A0A0L0BPG1 A0A0A1WV88 A0A0A1X6Q7 A0A0A1XSK0 A0A1D2N8Z7 A0A2H8TPE2 A0A2S2NWB1 A0A034WHY5 A0A1B6FJ75 A0A1I8MPK0 C4WTG8 A0A0K8VYX5 A0A1B6K362 A0A1I8PFW9 A0A1B6LL22 A0A0P4VZF1 A0A1A9WIH7 A0A1B0A1Z4
A0A1B0C9D2 A0A0K8TSW3 A0A2P8XZ43 A0A1B0DC51 A0A2J7QIP8 A0A067RNI5 A0A084WQZ4 A0A182H1Z1 T1DNV5 F4X6H1 A0A195E6W3 A0A310SA73 E2AM01 W5JWL5 Q1HQR7 A0A158NFZ7 Q16ME3 A0A026W6M9 A0A023F9P8 A0A161M4X0 A0A182INH1 D6WC51 A0A151IQD6 A0A1Q3G468 A0A2M4ASA2 A0A2M4BZB6 A0A182FKE5 A0A182KKD9 Q5TVQ1 A0A182HVU9 A0A224XXE8 A0A336M5R9 E2B3L0 V9IBB4 A0A069DXC2 A0A182WUM6 A0A1Q3G428 A0A2M3ZJI1 A0A1Q3G431 A0A182MB79 A0A182VDL3 A0A182U3S9 A0A0V0G5E1 A0A2A3EED4 V9ID41 A0A0B4J2N8 A0A182YKP1 A0A182PRN0 A0A1L8DVL3 A0A2J7RL08 A0A151XBY2 A0A182QWZ0 A0A195B2T2 A0A182WFV1 A0A1L8DVJ2 E9J609 U5EJY9 A0A0L7QKJ4 A0A0P4VRX9 R4G889 A0A2R7WBD9 A0A1W4XF77 A0A182R2X4 A0A182K195 A0A182NA36 E0V908 A0A0T6BD64 A0A1Y1MZ66 A0A1J1J188 A0A154PHX7 N6TTW3 W8BQJ2 A0A194PQN0 A0A1B6E3G8 A0A0L0BPG1 A0A0A1WV88 A0A0A1X6Q7 A0A0A1XSK0 A0A1D2N8Z7 A0A2H8TPE2 A0A2S2NWB1 A0A034WHY5 A0A1B6FJ75 A0A1I8MPK0 C4WTG8 A0A0K8VYX5 A0A1B6K362 A0A1I8PFW9 A0A1B6LL22 A0A0P4VZF1 A0A1A9WIH7 A0A1B0A1Z4
PDB
6I3V
E-value=2.72872e-34,
Score=360
Ontologies
PANTHER
Topology
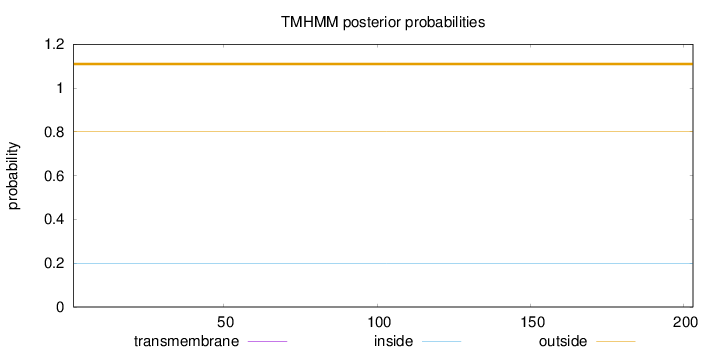
Length:
203
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00291
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.19852
outside
1 - 203
Population Genetic Test Statistics
Pi
279.117618
Theta
171.540038
Tajima's D
1.706216
CLR
123.183319
CSRT
0.829008549572521
Interpretation
Uncertain
