Gene
KWMTBOMO15427
Pre Gene Modal
BGIBMGA004774
Annotation
PREDICTED:_translation_initiation_factor_IF-2?_mitochondrial_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.772 Mitochondrial Reliability : 2.121
Sequence
CDS
ATGTATAGAAATCTGCTCTTTAACAGGCTTCCGGTAAAATTATTACACAGGGAGTACCAGCTACCAAAAGTCAGCTGTTTTATAAAAGCACTTACAAATAATAATTTGAAAGTTAGACGAATTTCTATATCTGTAGTATGCTGGAAGAAAAGACGATCGGAGGAAGAAAAGGCAGCAGTAAGTCAGGGTAGATCGGCAATCAGATATTCTCCAAAAGCTGGTTCAGAGGTTGTAGGAATTTGGAGGAACATGACAGTACAAGACGTTGCTGATGTGCTAAAAAAGGATATTGACCATGTTTTTAAAGTGCTAAATTTTGCTGACAAAACTGGAATGACATACACAAAGGATACAGTCATACACAATCCACAGGTGGTCAGGGACATTGTCAAAATATCAGGATGCAGATTTAAGTTGGTAGCAAGACCTGATGCTGAAATGAAAGATGAGGAGAACAAAGACGCAGTCCCGGCGCCTCCTGCGGCCGAGGAGGCGCTTCGGCCTCGACCTCCCGTGGTGTGTGTGATGGGGCATGTGGACCACGGAAAAACTACTCTACTAGATGCTCTCAGGGACACAGCGGTAGTGGAAACAGAGTTTGGGGGTATCACTCAGCATATTGGAGCTTTTGAGGTGAAACTGCCCAGTGGTGGGAAGGTGACATTCCTGGACACTCCCGGACACGCGGCCTTCACGTCCATGAGGAGTCGCGGCGCGCACTCCACCGACATCGTGGTGCTGGTCGTCGCCGCTGACGATGGTGTCATGGAACAAACACTCGAGTCCATTCGAATGGCGAGAGAAGCTAATGTTCCTATATTGGTGGCCATCAACAAAATCGACAAACCAGGTGTGAACGTTGAGCGCGTGTTGCGCGGGCTGGCCCAGCACGGCGTGGTGTGCGAGCAGGCGGGCGGCGAGGTGCAGGCCGTGCCCGTGTCGGCGCTGCGCCGCCGGAACCTCGACGCGCTGCTCGAGGCCCTCACCCTGCAGGCGCAGCTCATGCAGCTCCGCGCCGACCCCGCCGGACAGGTCGAGGGCGTCGTCATTGAAGCACTGGTGGACCCTAGGACCGGCAAGCAGGCGACGGTGCTGGTGCGGCGCGGGACGCTGCGGCGCGGCTGCACGCTGGTGGCGGGCGCGGGCTGGGCCAAGGTTCGCATTTTGAGGTCGGCGGAGGGTGCGTCCATAGAGGAGGCGCCCCCCTCCTGCCCCGTGGTGGTGCTCGGCTGGAGAGAGTTGCCCGCCGCCGGGGACCTGGTGCTCGAGGTCGCCTCTGAGAAGCGCGCGGCGGAGGTGATGCGGTGGCGCGCGGCGCAGCGCATGAAGGAGAAGCAGCAGTCCGACTCGGAAGTTATCGAAGCCAAGATGTCGGAGCACCAGCAGCGGTACCGGGCCATGCTGTCCCGGAAGCGGGCGATGGGCCGCTACAAACTGAAGCCGGAAGGTCCGCGGGAGAAAATGATCAAGGAGGACACGCATCCCGCGCTCAACGTCATCGTCAAAGGCGACGTGGATGGTTCAGTGGAGGCCATCCTGGACATCCTGGACACGTACGACGACCACGAGCGGGTCCGACTCGATCTCGTGCACTTTGGCGTGGGACAGGTCACGCCCAACGACCTGGAGATAGCGGAAGCCTTCAACGCTATAATATACGCGTTCAACGTGGAGTGCCCGCCTGCGGTCGCGGCGGACGCCAAGGACCTCAACATCTCCGTGAGGAAACACAACGTCATATACCGGCTGGTGGACGACGTCAAGCAGGAGATCACCGCCAGGATACCGGTCACGCAGACCGAGGAAGTCATCGGCGAAGCCAACGTGCTGCAGGAGTTCATGGTGTCGGAGGGCAGGAGGAAGGTGCCGGTGGCGGGCTGCCGCTGCGTGCGAGGCTCGCTGGCCAAGAACGCGCTCTACAAGATCATCAGGGGACAGGAAACCGTCTTCGAAGGTAAACTGGTCTCGATGAAGCACCTGAAGGAGGAGGTGCCGACGATAAAGCGCGACCAGGAGTGCGGCCTGAAGTTCGACGACCCCGAGGCGCGCGTGCAGCCCGGCGACACCGTGCTGTGCTACCGCCTGCGGGACGTCGCGCCGCGCACCGACTGGGACCCCGGCTTCTAG
Protein
MYRNLLFNRLPVKLLHREYQLPKVSCFIKALTNNNLKVRRISISVVCWKKRRSEEEKAAVSQGRSAIRYSPKAGSEVVGIWRNMTVQDVADVLKKDIDHVFKVLNFADKTGMTYTKDTVIHNPQVVRDIVKISGCRFKLVARPDAEMKDEENKDAVPAPPAAEEALRPRPPVVCVMGHVDHGKTTLLDALRDTAVVETEFGGITQHIGAFEVKLPSGGKVTFLDTPGHAAFTSMRSRGAHSTDIVVLVVAADDGVMEQTLESIRMAREANVPILVAINKIDKPGVNVERVLRGLAQHGVVCEQAGGEVQAVPVSALRRRNLDALLEALTLQAQLMQLRADPAGQVEGVVIEALVDPRTGKQATVLVRRGTLRRGCTLVAGAGWAKVRILRSAEGASIEEAPPSCPVVVLGWRELPAAGDLVLEVASEKRAAEVMRWRAAQRMKEKQQSDSEVIEAKMSEHQQRYRAMLSRKRAMGRYKLKPEGPREKMIKEDTHPALNVIVKGDVDGSVEAILDILDTYDDHERVRLDLVHFGVGQVTPNDLEIAEAFNAIIYAFNVECPPAVAADAKDLNISVRKHNVIYRLVDDVKQEITARIPVTQTEEVIGEANVLQEFMVSEGRRKVPVAGCRCVRGSLAKNALYKIIRGQETVFEGKLVSMKHLKEEVPTIKRDQECGLKFDDPEARVQPGDTVLCYRLRDVAPRTDWDPGF
Summary
Uniprot
H9J5I4
A0A2A4JYC4
A0A194PVW3
A0A194RIP9
A0A2H1V9T8
A0A212FBY8
+ More
A0A3S2LJL8 A0A067RTM6 A0A2J7RPR8 A0A1B6EDF4 A0A0A9WIR4 D6WQE9 A0A2R7WB56 A0A0P6BZI4 A0A0P5NZV9 A0A164PVX7 A0A0P6IVZ5 A0A0P5P6S6 A0A131XE58 A0A224ZA04 G3MKY6 A0A0P4W2N6 A0A131YWF1 E9H0D1 T1HV46 L7LUV5 A0A0N8B015 A0A023F1Z1 A0A0V0G8W4 K7J222 V5IG97 E2A3Z7 A0A232EH32 A0A182QTB3 A0A182MQ52 A0A3L8DR39 A0A182VZH2 A0A026WBJ9 A0A151X0H0 A0A182J657 A0A084WBR2 A0A0M4ELH8 A0A182NRZ3 A0A1Q3F4Y9 B4IH13 A0A0P5MLH1 A0A182FQD2 A0A182PGI6 B3P5S5 W5JIN4 Q9VAV2 A0A1I8N1A4 A0A151JS15 Q8MS23 A0A0C9RE90 B4K6Z7 E2BK40 B3LV81 B0W482 A0A0A1XTD1 A0A182RZ88 A0A1W4UG05 A0A195C3V8 A0A1I8P923 A0A1W4XKQ0 F4W750 A0A1D1W5Y5 A0A1A9YHZ6 U4UJ27 A0A195EYK8 N6TJT9 A0A182HCL2 B4QYR6 A0A182KFB4 A0A1S4F479 Q17G92 B4PQS3 B4MC51 A0A0J7KZZ9 B4JGP8 A0A336MIR5 A0A0K8VDJ2 A0A0P5G3A9 A0A0P5CIB8 A0A0P5UMV4 A0A1A9ZFP7 A0A0P5CXN3 A0A0Q9XC67 A0A1A9X2L9 A0A1A9UNN4 A0A182VAG0 A0A182WSM9 A0A3B0JHV5 A0A2H8TZA3 A0A0P8YG93 A0A182TMZ6 A0A182HLI0 Q7PKU6 B4N8N7
A0A3S2LJL8 A0A067RTM6 A0A2J7RPR8 A0A1B6EDF4 A0A0A9WIR4 D6WQE9 A0A2R7WB56 A0A0P6BZI4 A0A0P5NZV9 A0A164PVX7 A0A0P6IVZ5 A0A0P5P6S6 A0A131XE58 A0A224ZA04 G3MKY6 A0A0P4W2N6 A0A131YWF1 E9H0D1 T1HV46 L7LUV5 A0A0N8B015 A0A023F1Z1 A0A0V0G8W4 K7J222 V5IG97 E2A3Z7 A0A232EH32 A0A182QTB3 A0A182MQ52 A0A3L8DR39 A0A182VZH2 A0A026WBJ9 A0A151X0H0 A0A182J657 A0A084WBR2 A0A0M4ELH8 A0A182NRZ3 A0A1Q3F4Y9 B4IH13 A0A0P5MLH1 A0A182FQD2 A0A182PGI6 B3P5S5 W5JIN4 Q9VAV2 A0A1I8N1A4 A0A151JS15 Q8MS23 A0A0C9RE90 B4K6Z7 E2BK40 B3LV81 B0W482 A0A0A1XTD1 A0A182RZ88 A0A1W4UG05 A0A195C3V8 A0A1I8P923 A0A1W4XKQ0 F4W750 A0A1D1W5Y5 A0A1A9YHZ6 U4UJ27 A0A195EYK8 N6TJT9 A0A182HCL2 B4QYR6 A0A182KFB4 A0A1S4F479 Q17G92 B4PQS3 B4MC51 A0A0J7KZZ9 B4JGP8 A0A336MIR5 A0A0K8VDJ2 A0A0P5G3A9 A0A0P5CIB8 A0A0P5UMV4 A0A1A9ZFP7 A0A0P5CXN3 A0A0Q9XC67 A0A1A9X2L9 A0A1A9UNN4 A0A182VAG0 A0A182WSM9 A0A3B0JHV5 A0A2H8TZA3 A0A0P8YG93 A0A182TMZ6 A0A182HLI0 Q7PKU6 B4N8N7
Pubmed
19121390
26354079
22118469
24845553
25401762
18362917
+ More
19820115 28049606 28797301 22216098 26830274 21292972 25576852 25474469 20075255 25765539 20798317 28648823 30249741 24508170 24438588 17994087 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25830018 21719571 27649274 23537049 26483478 17510324 17550304 12364791 14747013 17210077
19820115 28049606 28797301 22216098 26830274 21292972 25576852 25474469 20075255 25765539 20798317 28648823 30249741 24508170 24438588 17994087 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25830018 21719571 27649274 23537049 26483478 17510324 17550304 12364791 14747013 17210077
EMBL
BABH01028205
NWSH01000383
PCG76809.1
KQ459596
KPI95270.1
KQ460118
+ More
KPJ17703.1 ODYU01001231 SOQ37162.1 AGBW02009250 OWR51259.1 RSAL01000001 RVE55305.1 KK852425 KDR24160.1 NEVH01001355 PNF42826.1 GEDC01001335 JAS35963.1 GBHO01038859 GBRD01001158 JAG04745.1 JAG64663.1 KQ971354 EFA07686.2 KK854565 PTY16922.1 GDIP01008125 JAM95590.1 GDIQ01142265 JAL09461.1 LRGB01002544 KZS07191.1 GDIQ01016895 JAN77842.1 GDIQ01133023 JAL18703.1 GEFH01003058 JAP65523.1 GFPF01012747 MAA23893.1 JO842537 AEO34154.1 GDRN01096834 JAI59267.1 GEDV01005675 JAP82882.1 GL732580 EFX74841.1 ACPB03003906 GACK01010420 JAA54614.1 GDIQ01226139 JAK25586.1 GBBI01003135 JAC15577.1 GECL01001529 JAP04595.1 GANP01007067 JAB77401.1 GL436519 EFN71849.1 NNAY01004622 OXU17644.1 AXCN02000256 AXCM01005462 AXCM01005463 AXCM01005464 QOIP01000005 RLU22905.1 KK107293 EZA53462.1 KQ982612 KYQ53846.1 ATLV01022425 KE525332 KFB47656.1 CP012526 ALC46152.1 GFDL01012414 JAV22631.1 CH480837 EDW49131.1 GDIQ01157664 JAK94061.1 CH954182 EDV53325.1 ADMH02001287 ETN63153.1 AE014297 BT150258 AAF56796.2 AGV77160.1 KQ978579 KYN30025.1 AY119138 AAM50998.1 GBYB01014903 GBYB01014904 JAG84670.1 JAG84671.1 CH933806 EDW15284.2 GL448740 EFN83948.1 CH902617 EDV43613.2 DS231835 EDS32926.1 GBXI01000052 JAD14240.1 KQ978317 KYM95295.1 GL887813 EGI69934.1 BDGG01000015 GAV07598.1 KB632326 ERL92423.1 KQ981920 KYN32992.1 APGK01035004 APGK01035005 KB740923 ENN78138.1 JXUM01033558 KQ560993 KXJ80094.1 CM000364 EDX14735.1 CH477265 EAT45556.1 CM000160 EDW98412.1 CH940656 EDW58672.1 LBMM01001641 KMQ95956.1 CH916369 EDV92652.1 UFQS01001179 UFQT01001179 SSX09535.1 SSX29351.1 GDHF01015699 JAI36615.1 GDIQ01247944 JAK03781.1 GDIP01169720 JAJ53682.1 GDIP01114433 JAL89281.1 GDIP01163979 JAJ59423.1 KRG01496.1 OUUW01000005 SPP80293.1 GFXV01007434 MBW19239.1 KPU80384.1 APCN01000904 AAAB01008987 EAA43132.4 CH964232 EDW81488.2
KPJ17703.1 ODYU01001231 SOQ37162.1 AGBW02009250 OWR51259.1 RSAL01000001 RVE55305.1 KK852425 KDR24160.1 NEVH01001355 PNF42826.1 GEDC01001335 JAS35963.1 GBHO01038859 GBRD01001158 JAG04745.1 JAG64663.1 KQ971354 EFA07686.2 KK854565 PTY16922.1 GDIP01008125 JAM95590.1 GDIQ01142265 JAL09461.1 LRGB01002544 KZS07191.1 GDIQ01016895 JAN77842.1 GDIQ01133023 JAL18703.1 GEFH01003058 JAP65523.1 GFPF01012747 MAA23893.1 JO842537 AEO34154.1 GDRN01096834 JAI59267.1 GEDV01005675 JAP82882.1 GL732580 EFX74841.1 ACPB03003906 GACK01010420 JAA54614.1 GDIQ01226139 JAK25586.1 GBBI01003135 JAC15577.1 GECL01001529 JAP04595.1 GANP01007067 JAB77401.1 GL436519 EFN71849.1 NNAY01004622 OXU17644.1 AXCN02000256 AXCM01005462 AXCM01005463 AXCM01005464 QOIP01000005 RLU22905.1 KK107293 EZA53462.1 KQ982612 KYQ53846.1 ATLV01022425 KE525332 KFB47656.1 CP012526 ALC46152.1 GFDL01012414 JAV22631.1 CH480837 EDW49131.1 GDIQ01157664 JAK94061.1 CH954182 EDV53325.1 ADMH02001287 ETN63153.1 AE014297 BT150258 AAF56796.2 AGV77160.1 KQ978579 KYN30025.1 AY119138 AAM50998.1 GBYB01014903 GBYB01014904 JAG84670.1 JAG84671.1 CH933806 EDW15284.2 GL448740 EFN83948.1 CH902617 EDV43613.2 DS231835 EDS32926.1 GBXI01000052 JAD14240.1 KQ978317 KYM95295.1 GL887813 EGI69934.1 BDGG01000015 GAV07598.1 KB632326 ERL92423.1 KQ981920 KYN32992.1 APGK01035004 APGK01035005 KB740923 ENN78138.1 JXUM01033558 KQ560993 KXJ80094.1 CM000364 EDX14735.1 CH477265 EAT45556.1 CM000160 EDW98412.1 CH940656 EDW58672.1 LBMM01001641 KMQ95956.1 CH916369 EDV92652.1 UFQS01001179 UFQT01001179 SSX09535.1 SSX29351.1 GDHF01015699 JAI36615.1 GDIQ01247944 JAK03781.1 GDIP01169720 JAJ53682.1 GDIP01114433 JAL89281.1 GDIP01163979 JAJ59423.1 KRG01496.1 OUUW01000005 SPP80293.1 GFXV01007434 MBW19239.1 KPU80384.1 APCN01000904 AAAB01008987 EAA43132.4 CH964232 EDW81488.2
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000027135 UP000235965 UP000007266 UP000076858 UP000000305 UP000015103 UP000002358 UP000000311 UP000215335 UP000075886 UP000075883 UP000279307 UP000075920 UP000053097 UP000075809 UP000075880 UP000030765 UP000092553 UP000075884 UP000001292 UP000069272 UP000075885 UP000008711 UP000000673 UP000000803 UP000095301 UP000078492 UP000009192 UP000008237 UP000007801 UP000002320 UP000075900 UP000192221 UP000078542 UP000095300 UP000192223 UP000007755 UP000186922 UP000092443 UP000030742 UP000078541 UP000019118 UP000069940 UP000249989 UP000000304 UP000075881 UP000008820 UP000002282 UP000008792 UP000036403 UP000001070 UP000092445 UP000091820 UP000078200 UP000075903 UP000076407 UP000268350 UP000075902 UP000075840 UP000007062 UP000007798
UP000027135 UP000235965 UP000007266 UP000076858 UP000000305 UP000015103 UP000002358 UP000000311 UP000215335 UP000075886 UP000075883 UP000279307 UP000075920 UP000053097 UP000075809 UP000075880 UP000030765 UP000092553 UP000075884 UP000001292 UP000069272 UP000075885 UP000008711 UP000000673 UP000000803 UP000095301 UP000078492 UP000009192 UP000008237 UP000007801 UP000002320 UP000075900 UP000192221 UP000078542 UP000095300 UP000192223 UP000007755 UP000186922 UP000092443 UP000030742 UP000078541 UP000019118 UP000069940 UP000249989 UP000000304 UP000075881 UP000008820 UP000002282 UP000008792 UP000036403 UP000001070 UP000092445 UP000091820 UP000078200 UP000075903 UP000076407 UP000268350 UP000075902 UP000075840 UP000007062 UP000007798
Interpro
Gene 3D
ProteinModelPortal
H9J5I4
A0A2A4JYC4
A0A194PVW3
A0A194RIP9
A0A2H1V9T8
A0A212FBY8
+ More
A0A3S2LJL8 A0A067RTM6 A0A2J7RPR8 A0A1B6EDF4 A0A0A9WIR4 D6WQE9 A0A2R7WB56 A0A0P6BZI4 A0A0P5NZV9 A0A164PVX7 A0A0P6IVZ5 A0A0P5P6S6 A0A131XE58 A0A224ZA04 G3MKY6 A0A0P4W2N6 A0A131YWF1 E9H0D1 T1HV46 L7LUV5 A0A0N8B015 A0A023F1Z1 A0A0V0G8W4 K7J222 V5IG97 E2A3Z7 A0A232EH32 A0A182QTB3 A0A182MQ52 A0A3L8DR39 A0A182VZH2 A0A026WBJ9 A0A151X0H0 A0A182J657 A0A084WBR2 A0A0M4ELH8 A0A182NRZ3 A0A1Q3F4Y9 B4IH13 A0A0P5MLH1 A0A182FQD2 A0A182PGI6 B3P5S5 W5JIN4 Q9VAV2 A0A1I8N1A4 A0A151JS15 Q8MS23 A0A0C9RE90 B4K6Z7 E2BK40 B3LV81 B0W482 A0A0A1XTD1 A0A182RZ88 A0A1W4UG05 A0A195C3V8 A0A1I8P923 A0A1W4XKQ0 F4W750 A0A1D1W5Y5 A0A1A9YHZ6 U4UJ27 A0A195EYK8 N6TJT9 A0A182HCL2 B4QYR6 A0A182KFB4 A0A1S4F479 Q17G92 B4PQS3 B4MC51 A0A0J7KZZ9 B4JGP8 A0A336MIR5 A0A0K8VDJ2 A0A0P5G3A9 A0A0P5CIB8 A0A0P5UMV4 A0A1A9ZFP7 A0A0P5CXN3 A0A0Q9XC67 A0A1A9X2L9 A0A1A9UNN4 A0A182VAG0 A0A182WSM9 A0A3B0JHV5 A0A2H8TZA3 A0A0P8YG93 A0A182TMZ6 A0A182HLI0 Q7PKU6 B4N8N7
A0A3S2LJL8 A0A067RTM6 A0A2J7RPR8 A0A1B6EDF4 A0A0A9WIR4 D6WQE9 A0A2R7WB56 A0A0P6BZI4 A0A0P5NZV9 A0A164PVX7 A0A0P6IVZ5 A0A0P5P6S6 A0A131XE58 A0A224ZA04 G3MKY6 A0A0P4W2N6 A0A131YWF1 E9H0D1 T1HV46 L7LUV5 A0A0N8B015 A0A023F1Z1 A0A0V0G8W4 K7J222 V5IG97 E2A3Z7 A0A232EH32 A0A182QTB3 A0A182MQ52 A0A3L8DR39 A0A182VZH2 A0A026WBJ9 A0A151X0H0 A0A182J657 A0A084WBR2 A0A0M4ELH8 A0A182NRZ3 A0A1Q3F4Y9 B4IH13 A0A0P5MLH1 A0A182FQD2 A0A182PGI6 B3P5S5 W5JIN4 Q9VAV2 A0A1I8N1A4 A0A151JS15 Q8MS23 A0A0C9RE90 B4K6Z7 E2BK40 B3LV81 B0W482 A0A0A1XTD1 A0A182RZ88 A0A1W4UG05 A0A195C3V8 A0A1I8P923 A0A1W4XKQ0 F4W750 A0A1D1W5Y5 A0A1A9YHZ6 U4UJ27 A0A195EYK8 N6TJT9 A0A182HCL2 B4QYR6 A0A182KFB4 A0A1S4F479 Q17G92 B4PQS3 B4MC51 A0A0J7KZZ9 B4JGP8 A0A336MIR5 A0A0K8VDJ2 A0A0P5G3A9 A0A0P5CIB8 A0A0P5UMV4 A0A1A9ZFP7 A0A0P5CXN3 A0A0Q9XC67 A0A1A9X2L9 A0A1A9UNN4 A0A182VAG0 A0A182WSM9 A0A3B0JHV5 A0A2H8TZA3 A0A0P8YG93 A0A182TMZ6 A0A182HLI0 Q7PKU6 B4N8N7
PDB
6GB2
E-value=7.81559e-152,
Score=1380
Ontologies
GO
PANTHER
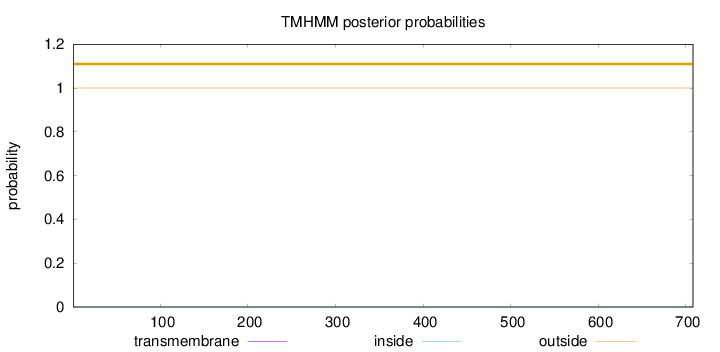
Topology
Length:
708
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00013
outside
1 - 708
Population Genetic Test Statistics
Pi
184.078075
Theta
153.601008
Tajima's D
0.395085
CLR
0.08346
CSRT
0.479426028698565
Interpretation
Uncertain
