Pre Gene Modal
BGIBMGA004741
Annotation
PREDICTED:_HIV_Tat-specific_factor_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.278
Sequence
CDS
ATGGCTAAACTAAATCCAAGAACGGGATTTGTGATCAAACTTTCCGAAGAGACAGTTCAAAAGTTAGCGGAAGAGGAAAAAGAAACAAAAGAAAAATCTACATATAATCAGATAGAGGATACTACATCGGTAGAAGATATTAATAATGACAAAATTCAAAATGTTCAAGATGTGAAAACAACATCACAAACTTTAGATGTACCTACTTCCGATTCCATAGAAAAATCTACTGTAGAAAGTACTCTGCAGAAAGAAAATTCATCACAAACTGAAAAAACAGTTCTGAACATAACTGATGATAAGGGTGAAAATGATAAGGATAATACACTTGAAGATAATGAAAAAACAAAGAAACCCTCCAAATCTATTACTGATTCTAGTACTTGGGGTGAGTATGCATCATACATAACTTACAAAGATAATGAAGCTGTATATACTGACCCCTCATCAGGTCAAGCTTACACTTGGAATAAAACCAGCAATGCTTGGGATGCTAAAGGTGATGAAGCACCTGGTCGTGTTTACAGCTATGAAGATGACACTCATATATACACAGAGGCTGATGGTTCAAAATATTTTTGGGATGAAACCAAGAAGGCTTGGATACCAAAAGTTGATGATGACTTTTTAGCTTATTATCAAATGTCTTATGGATTTATTGAAAATAAGAAAGATGATGATGAGAAAGAGAAAGGTAGTGACAAGAAGAAGGAGTTGCAAAAAGAAGCTATGGCTGTCAAAAGGAAAAGTGAACCGCAGTGGTTTGAACCTACTGATGATAAAAACACCAAAGTTTATGTATCAAACTTACCTCTAGACATGACTGAACAAGAATTTGTGGACTTAATGCAGAAGTGTGGTCTCGTTGAACGAGATCCAAGTAACCAAAAAATGAAAGTTAAATTATACATGGACAAAGAACAAAATTGTTTCAAGGGTGATGCATTATGTACTTATATAAAAATAGAATCCGTAGATTTAGCATTAAAATTGTTGGATGGCAGCGATTGTAAAGGCAATACCATTAAAGTGGAAAGAGCTCAATTTCAACTAAAAGGGGAATACAATCCTGCCCTCAAACCTAAGAAAAGAAGAAAAAAAGAGCTGGAAAAAATTAAAAAAATGAAAGAAAAACTATTTGATTGGAGGCCAGAGAAGTTTATTGGTGAACGTTCTAAACACGAAAGAGTAGTTATTGTTAAAAATTTATTCCACTCTAGTGATTTTGATAAAGAGGTTCAGCTTATTCTGGACTATCAGCAAGATTTGAGGGAAGAATGTGGAAAATGTGGAGACGTTCGGAAAGTAGTGCTCTATGATCGCCATCCTGAAGGAGTGGCACAGATAACTATGAAAGAGCCGGAACAAGCTGATGCTGTTGTTCAGCTGATCAACGGAAGATGGTTTGGCAAAAGACAAATCACTGCAGAGATATGGGATGGCAAAACTAAGTACAAAATTGCTGAAACTGATGCAGACATTAATCAGAGGTTAGATAAATGGGGTAAATTTTTGGAAGAAAAAGAAGATGATAATAAGGTAATTAAATTGCCTGTTGAAGAAAAAGACACAGATGAGCTTAAAGTTGAAGAGGTTTCTATTGAAAAAGAAAATACTCAATACCTATCCGAAAAAAGTGATATAAACAATGTAAAATTAGAAATAAATCAATGA
Protein
MAKLNPRTGFVIKLSEETVQKLAEEEKETKEKSTYNQIEDTTSVEDINNDKIQNVQDVKTTSQTLDVPTSDSIEKSTVESTLQKENSSQTEKTVLNITDDKGENDKDNTLEDNEKTKKPSKSITDSSTWGEYASYITYKDNEAVYTDPSSGQAYTWNKTSNAWDAKGDEAPGRVYSYEDDTHIYTEADGSKYFWDETKKAWIPKVDDDFLAYYQMSYGFIENKKDDDEKEKGSDKKKELQKEAMAVKRKSEPQWFEPTDDKNTKVYVSNLPLDMTEQEFVDLMQKCGLVERDPSNQKMKVKLYMDKEQNCFKGDALCTYIKIESVDLALKLLDGSDCKGNTIKVERAQFQLKGEYNPALKPKKRRKKELEKIKKMKEKLFDWRPEKFIGERSKHERVVIVKNLFHSSDFDKEVQLILDYQQDLREECGKCGDVRKVVLYDRHPEGVAQITMKEPEQADAVVQLINGRWFGKRQITAEIWDGKTKYKIAETDADINQRLDKWGKFLEEKEDDNKVIKLPVEEKDTDELKVEEVSIEKENTQYLSEKSDINNVKLEINQ
Summary
Uniprot
H9J5F1
A0A2H1VRL5
A0A212FC14
A0A194RK42
A0A0L7KT78
A0A2A4JXR8
+ More
A0A194PQM5 A0A1Y1KY94 A0A1W4WYW9 D7EJG5 A0A0C9RRA1 A0A1Q3G184 A0A1Q3G175 A0A2M4ANZ5 A0A2M4CS54 A0A2M4AM76 A0A2M4AMR7 A0A0P6IUZ6 A0A2M4CTF3 W8B1F8 A0A1B6CZN8 B3NE82 A0A026WQ05 A0A195F576 A0A158NJW1 B4PE28 A0A1W4UWR4 A0A0A1XAC0 A0A0A9XG48 A0A195DV91 A0A0K8SSD9 A0A146LBP2 A0A1A9V3T3 A0A1A9Y0W1 Q2LYN4 A0A3B0JT67 A0A0K8U5W6 A0A0J9S0R5 A0A034VCD8 A0A1A9W771 A0A2A3EMB6 Q7K204 A0A1S4ESR1 A0A088AKE1 B3M8J4 A0A1L8DGF0 A0A1B0B6V4 A0A232F3D2 A0A1J1I7E8 K7JAK9 A0A3B0KNB4 A0A2S2PRV1 B4KWT8 A0A336M4B4 B4IXC1 A0A2H8TT29 A0A1I8PBJ2
A0A194PQM5 A0A1Y1KY94 A0A1W4WYW9 D7EJG5 A0A0C9RRA1 A0A1Q3G184 A0A1Q3G175 A0A2M4ANZ5 A0A2M4CS54 A0A2M4AM76 A0A2M4AMR7 A0A0P6IUZ6 A0A2M4CTF3 W8B1F8 A0A1B6CZN8 B3NE82 A0A026WQ05 A0A195F576 A0A158NJW1 B4PE28 A0A1W4UWR4 A0A0A1XAC0 A0A0A9XG48 A0A195DV91 A0A0K8SSD9 A0A146LBP2 A0A1A9V3T3 A0A1A9Y0W1 Q2LYN4 A0A3B0JT67 A0A0K8U5W6 A0A0J9S0R5 A0A034VCD8 A0A1A9W771 A0A2A3EMB6 Q7K204 A0A1S4ESR1 A0A088AKE1 B3M8J4 A0A1L8DGF0 A0A1B0B6V4 A0A232F3D2 A0A1J1I7E8 K7JAK9 A0A3B0KNB4 A0A2S2PRV1 B4KWT8 A0A336M4B4 B4IXC1 A0A2H8TT29 A0A1I8PBJ2
Pubmed
19121390
22118469
26354079
26227816
28004739
18362917
+ More
19820115 26999592 24495485 17994087 18057021 24508170 30249741 21347285 17550304 25830018 25401762 26823975 15632085 22936249 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255
19820115 26999592 24495485 17994087 18057021 24508170 30249741 21347285 17550304 25830018 25401762 26823975 15632085 22936249 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255
EMBL
BABH01028207
ODYU01004030
SOQ43491.1
AGBW02009250
OWR51257.1
KQ460118
+ More
KPJ17705.1 JTDY01006137 KOB66256.1 NWSH01000383 PCG76811.1 KQ459596 KPI95268.1 GEZM01072553 JAV65471.1 DS497704 EFA12731.1 GBYB01011070 JAG80837.1 GFDL01001481 JAV33564.1 GFDL01001486 JAV33559.1 GGFK01009156 MBW42477.1 GGFL01003931 MBW68109.1 GGFK01008550 MBW41871.1 GGFK01008577 MBW41898.1 GDUN01000945 JAN94974.1 GGFL01003930 MBW68108.1 GAMC01015702 GAMC01015701 JAB90853.1 GEDC01018371 JAS18927.1 CH954178 EDV52646.1 KQS44367.1 KK107135 QOIP01000006 EZA58058.1 RLU21927.1 KQ981820 KYN35327.1 ADTU01018410 ADTU01018411 ADTU01018412 CM000159 EDW95041.1 KRK02205.1 GBXI01006048 JAD08244.1 GBHO01027544 JAG16060.1 KQ980304 KYN16766.1 GBRD01009745 JAG56079.1 GDHC01017656 GDHC01013530 GDHC01008722 JAQ00973.1 JAQ05099.1 JAQ09907.1 CH379069 EAL29828.2 OUUW01000009 SPP85307.1 GDHF01030270 GDHF01015293 JAI22044.1 JAI37021.1 CM002912 KMZ00995.1 GAKP01017971 JAC40981.1 KZ288217 PBC32422.1 AY061345 AE014296 AAL28893.1 AAN12166.1 AAN12167.1 CH902618 EDV39968.2 GFDF01008670 JAV05414.1 JXJN01009246 JXJN01009247 NNAY01001048 OXU25346.1 CVRI01000043 CRK96099.1 AAZX01002446 AAZX01002448 AAZX01009950 AAZX01015712 AAZX01018407 SPP85308.1 GGMR01019439 MBY32058.1 CH933809 EDW18559.2 UFQT01000381 SSX23779.1 CH916366 EDV97453.1 GFXV01005581 MBW17386.1
KPJ17705.1 JTDY01006137 KOB66256.1 NWSH01000383 PCG76811.1 KQ459596 KPI95268.1 GEZM01072553 JAV65471.1 DS497704 EFA12731.1 GBYB01011070 JAG80837.1 GFDL01001481 JAV33564.1 GFDL01001486 JAV33559.1 GGFK01009156 MBW42477.1 GGFL01003931 MBW68109.1 GGFK01008550 MBW41871.1 GGFK01008577 MBW41898.1 GDUN01000945 JAN94974.1 GGFL01003930 MBW68108.1 GAMC01015702 GAMC01015701 JAB90853.1 GEDC01018371 JAS18927.1 CH954178 EDV52646.1 KQS44367.1 KK107135 QOIP01000006 EZA58058.1 RLU21927.1 KQ981820 KYN35327.1 ADTU01018410 ADTU01018411 ADTU01018412 CM000159 EDW95041.1 KRK02205.1 GBXI01006048 JAD08244.1 GBHO01027544 JAG16060.1 KQ980304 KYN16766.1 GBRD01009745 JAG56079.1 GDHC01017656 GDHC01013530 GDHC01008722 JAQ00973.1 JAQ05099.1 JAQ09907.1 CH379069 EAL29828.2 OUUW01000009 SPP85307.1 GDHF01030270 GDHF01015293 JAI22044.1 JAI37021.1 CM002912 KMZ00995.1 GAKP01017971 JAC40981.1 KZ288217 PBC32422.1 AY061345 AE014296 AAL28893.1 AAN12166.1 AAN12167.1 CH902618 EDV39968.2 GFDF01008670 JAV05414.1 JXJN01009246 JXJN01009247 NNAY01001048 OXU25346.1 CVRI01000043 CRK96099.1 AAZX01002446 AAZX01002448 AAZX01009950 AAZX01015712 AAZX01018407 SPP85308.1 GGMR01019439 MBY32058.1 CH933809 EDW18559.2 UFQT01000381 SSX23779.1 CH916366 EDV97453.1 GFXV01005581 MBW17386.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000218220
UP000053268
+ More
UP000192223 UP000007266 UP000008711 UP000053097 UP000279307 UP000078541 UP000005205 UP000002282 UP000192221 UP000078492 UP000078200 UP000092443 UP000001819 UP000268350 UP000091820 UP000242457 UP000000803 UP000079169 UP000005203 UP000007801 UP000092460 UP000215335 UP000183832 UP000002358 UP000009192 UP000001070 UP000095300
UP000192223 UP000007266 UP000008711 UP000053097 UP000279307 UP000078541 UP000005205 UP000002282 UP000192221 UP000078492 UP000078200 UP000092443 UP000001819 UP000268350 UP000091820 UP000242457 UP000000803 UP000079169 UP000005203 UP000007801 UP000092460 UP000215335 UP000183832 UP000002358 UP000009192 UP000001070 UP000095300
Pfam
PF00076 RRM_1
Interpro
Gene 3D
ProteinModelPortal
H9J5F1
A0A2H1VRL5
A0A212FC14
A0A194RK42
A0A0L7KT78
A0A2A4JXR8
+ More
A0A194PQM5 A0A1Y1KY94 A0A1W4WYW9 D7EJG5 A0A0C9RRA1 A0A1Q3G184 A0A1Q3G175 A0A2M4ANZ5 A0A2M4CS54 A0A2M4AM76 A0A2M4AMR7 A0A0P6IUZ6 A0A2M4CTF3 W8B1F8 A0A1B6CZN8 B3NE82 A0A026WQ05 A0A195F576 A0A158NJW1 B4PE28 A0A1W4UWR4 A0A0A1XAC0 A0A0A9XG48 A0A195DV91 A0A0K8SSD9 A0A146LBP2 A0A1A9V3T3 A0A1A9Y0W1 Q2LYN4 A0A3B0JT67 A0A0K8U5W6 A0A0J9S0R5 A0A034VCD8 A0A1A9W771 A0A2A3EMB6 Q7K204 A0A1S4ESR1 A0A088AKE1 B3M8J4 A0A1L8DGF0 A0A1B0B6V4 A0A232F3D2 A0A1J1I7E8 K7JAK9 A0A3B0KNB4 A0A2S2PRV1 B4KWT8 A0A336M4B4 B4IXC1 A0A2H8TT29 A0A1I8PBJ2
A0A194PQM5 A0A1Y1KY94 A0A1W4WYW9 D7EJG5 A0A0C9RRA1 A0A1Q3G184 A0A1Q3G175 A0A2M4ANZ5 A0A2M4CS54 A0A2M4AM76 A0A2M4AMR7 A0A0P6IUZ6 A0A2M4CTF3 W8B1F8 A0A1B6CZN8 B3NE82 A0A026WQ05 A0A195F576 A0A158NJW1 B4PE28 A0A1W4UWR4 A0A0A1XAC0 A0A0A9XG48 A0A195DV91 A0A0K8SSD9 A0A146LBP2 A0A1A9V3T3 A0A1A9Y0W1 Q2LYN4 A0A3B0JT67 A0A0K8U5W6 A0A0J9S0R5 A0A034VCD8 A0A1A9W771 A0A2A3EMB6 Q7K204 A0A1S4ESR1 A0A088AKE1 B3M8J4 A0A1L8DGF0 A0A1B0B6V4 A0A232F3D2 A0A1J1I7E8 K7JAK9 A0A3B0KNB4 A0A2S2PRV1 B4KWT8 A0A336M4B4 B4IXC1 A0A2H8TT29 A0A1I8PBJ2
PDB
2DIT
E-value=2.97921e-27,
Score=305
Ontologies
GO
PANTHER
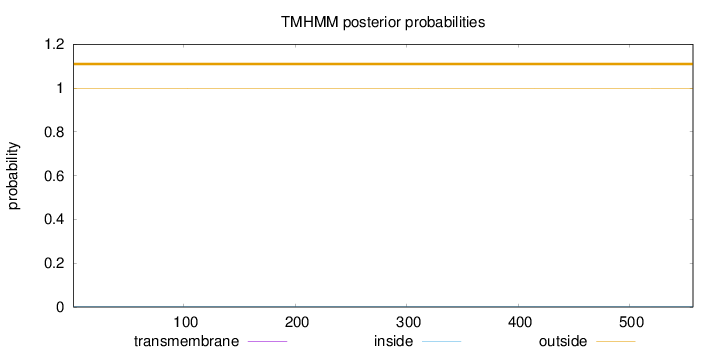
Topology
Length:
557
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00091
outside
1 - 557
Population Genetic Test Statistics
Pi
150.515677
Theta
196.503049
Tajima's D
-0.766382
CLR
34.04941
CSRT
0.185440727963602
Interpretation
Uncertain
