Gene
KWMTBOMO15424 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004775
Annotation
PREDICTED:_uridine_diphosphate_glucose_pyrophosphatase-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 3.164
Sequence
CDS
ATGGAAGATTTAAAGGATGTGTATTTGAGTCCATTACCGGAATCGCCGTACGTGAAGCCGTTCAGGTTCAATTACACGCAGAACGGCAAAGAGAAAACTTGGGATCTTCTAGAAGTTCATGACTCCGTTGCCATCATAGTATTTAATGTAACTAGAAGAGTAATGATATTCGTAAAACAATTTAGACCAGCCATTTATTACAATTCAATAAAACCAGAAGATAGAAAAGCATCAAAGATAGACACAAATAAGTATCCTGCATCTCTTGGTGTCGCTTTGGAAATATGTGCTGGAATCATTGACAAGAATTTACCCATAGAGGAAATTGCCAAGGAGGAGGTGCTTGAAGAGTGTGGCTACAATGTTGAGCTTTCAAATTTGGAAAAAGTTATAGCATATAGGTCAGGTGTCGGGGTTCAGGGTGCCTTACAAACAATGTTTTATTGTGAGGTCACAGATGATATGAAAACTGAACAGGGTGGCGGCGTTGACGATGAAATTATAGAAGTTATAGAGAAGACAATACCTGAAGTGGAAGAGATGCTTCGTTCACAAGACATCCTGTCCAGTCCACCGAGCTGTCTTTTTGCTTTAATGTGGTTTATACACAATAAAGCCGATAAGTTTAGGAAGACATTCGACCGAACTTGCGGGGAATGTAAGAAATCGAACAAATAA
Protein
MEDLKDVYLSPLPESPYVKPFRFNYTQNGKEKTWDLLEVHDSVAIIVFNVTRRVMIFVKQFRPAIYYNSIKPEDRKASKIDTNKYPASLGVALEICAGIIDKNLPIEEIAKEEVLEECGYNVELSNLEKVIAYRSGVGVQGALQTMFYCEVTDDMKTEQGGGVDDEIIEVIEKTIPEVEEMLRSQDILSSPPSCLFALMWFIHNKADKFRKTFDRTCGECKKSNK
Summary
Uniprot
H9J5I5
A0A3S2NSJ3
A0A212FBY6
I4DKY0
S4NNF5
A0A2H1VRP4
+ More
A0A194RNW1 D2A0B4 A0A336MPJ8 A0A336M6L8 T1DQ41 A0A336MRY2 A0A336KL19 A0A0K8WB32 A0A034W3H9 A0A1W7R5R8 A0A2M3ZAX5 A0A182FPU7 A0A0K8TQX5 A0A1S4EXV6 A0A2M3ZB57 A0A2M3ZJ31 A0A2M3ZJD1 W5JMR1 A0A2M4BY66 A0A1L8EEK4 A0A1L8EE70 A0A2M4BYD4 A0A182K1D8 Q17M97 A0A1L8EEF5 A0A182GYQ6 T1PA38 A0A2M4ALX7 B4M0A9 N6UDF7 A0A1I8Q3X0 A0A0L0BMX2 A0A182N5S7 A0A182XJ27 A0A182I7T2 A0A1Y1MNL6 A0A182JMI3 A0A0M3QXS9 W8C3Y4 Q7QB65 A0A182P1Q4 A0A1Q3FJC4 A0A182TRF0 A0A182V9H9 A0A182YGM5 B0WQD2 Q9VB64 A0A084WTK6 A0A1W4UIY6 B4PRT9 F2FB73 A0A240SX77 A0A1A9YSU7 A0A1B0B2P8 A0A182W1I8 A0A182RYM8 A0A0Q9X1U9 B4JRR5 A0A182L7C9 A0A3B0K3N3 A0A182QGF8 A0A1A9VFZ6 B3MT40 B5DWT8 T1JH04 B4IC58 B4QXF6 A0A1B0CQZ0 B4NBF6 B3P841 U5EWZ5 A0A1A9WF40 A0A1A9WR22 A0A1W4XS49 A0A2R7WYD3 A0A1B6GMR0 A0A1B0DBG7 A0A1L8DCN6 A0A1B0BK86 A0A1B0FCU4 V5GLU0 B4G2H4 A0A1B6M416 A0A1B6F258 A0A0N8AV12 A0A0N8EHD3 A0A164Z5B6 A0A067RHA2 A0A293N3C1 A0A1B6KJQ1 A0A0P4ZNX9 A0A0P5A5X9 A0A0P6AHW7
A0A194RNW1 D2A0B4 A0A336MPJ8 A0A336M6L8 T1DQ41 A0A336MRY2 A0A336KL19 A0A0K8WB32 A0A034W3H9 A0A1W7R5R8 A0A2M3ZAX5 A0A182FPU7 A0A0K8TQX5 A0A1S4EXV6 A0A2M3ZB57 A0A2M3ZJ31 A0A2M3ZJD1 W5JMR1 A0A2M4BY66 A0A1L8EEK4 A0A1L8EE70 A0A2M4BYD4 A0A182K1D8 Q17M97 A0A1L8EEF5 A0A182GYQ6 T1PA38 A0A2M4ALX7 B4M0A9 N6UDF7 A0A1I8Q3X0 A0A0L0BMX2 A0A182N5S7 A0A182XJ27 A0A182I7T2 A0A1Y1MNL6 A0A182JMI3 A0A0M3QXS9 W8C3Y4 Q7QB65 A0A182P1Q4 A0A1Q3FJC4 A0A182TRF0 A0A182V9H9 A0A182YGM5 B0WQD2 Q9VB64 A0A084WTK6 A0A1W4UIY6 B4PRT9 F2FB73 A0A240SX77 A0A1A9YSU7 A0A1B0B2P8 A0A182W1I8 A0A182RYM8 A0A0Q9X1U9 B4JRR5 A0A182L7C9 A0A3B0K3N3 A0A182QGF8 A0A1A9VFZ6 B3MT40 B5DWT8 T1JH04 B4IC58 B4QXF6 A0A1B0CQZ0 B4NBF6 B3P841 U5EWZ5 A0A1A9WF40 A0A1A9WR22 A0A1W4XS49 A0A2R7WYD3 A0A1B6GMR0 A0A1B0DBG7 A0A1L8DCN6 A0A1B0BK86 A0A1B0FCU4 V5GLU0 B4G2H4 A0A1B6M416 A0A1B6F258 A0A0N8AV12 A0A0N8EHD3 A0A164Z5B6 A0A067RHA2 A0A293N3C1 A0A1B6KJQ1 A0A0P4ZNX9 A0A0P5A5X9 A0A0P6AHW7
Pubmed
19121390
22118469
22651552
26354079
23622113
18362917
+ More
19820115 25348373 26369729 20920257 23761445 17510324 26483478 25315136 17994087 23537049 26108605 28004739 24495485 12364791 14747013 17210077 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 17550304 20966253 15632085 24845553
19820115 25348373 26369729 20920257 23761445 17510324 26483478 25315136 17994087 23537049 26108605 28004739 24495485 12364791 14747013 17210077 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 17550304 20966253 15632085 24845553
EMBL
BABH01028207
RSAL01000001
RVE55276.1
AGBW02009250
OWR51256.1
AK401948
+ More
KQ459596 BAM18570.1 KPI95267.1 GAIX01013951 JAA78609.1 ODYU01004030 SOQ43490.1 KQ460118 KPJ17706.1 KQ971338 EFA01723.1 UFQT01001990 SSX32322.1 UFQS01000630 UFQT01000630 SSX05547.1 SSX25906.1 GAMD01002789 JAA98801.1 SSX32321.1 SSX05546.1 SSX25905.1 GDHF01004037 JAI48277.1 GAKP01009703 GAKP01009702 GAKP01009701 GAKP01009700 JAC49249.1 GEHC01001290 JAV46355.1 GGFM01004945 MBW25696.1 GDAI01001055 JAI16548.1 GGFM01005008 MBW25759.1 GGFM01007724 MBW28475.1 GGFM01007797 MBW28548.1 ADMH02000944 ETN64603.1 GGFJ01008885 MBW58026.1 GFDG01001806 JAV16993.1 GFDG01001801 JAV16998.1 GGFJ01008883 MBW58024.1 CH477207 EAT47852.1 GFDG01001820 JAV16979.1 JXUM01021495 KQ560602 KXJ81678.1 KA645554 AFP60183.1 GGFK01008453 MBW41774.1 CH940650 EDW67271.1 APGK01026955 APGK01026956 KB740648 KB631780 ENN79735.1 ERL86037.1 JRES01001623 KNC21450.1 APCN01002552 GEZM01026068 GEZM01026067 JAV87264.1 CP012526 ALC46399.1 GAMC01009912 GAMC01009911 JAB96644.1 AAAB01008880 EAA08524.4 GFDL01007420 JAV27625.1 DS232039 EDS32803.1 AE014297 AAF56679.3 ALI30643.1 ATLV01026898 KE525420 KFB53550.1 CM000160 EDW98532.2 BT126160 ADZ99412.1 JXJN01007687 CH933806 KRG02070.1 CH916373 EDV94455.1 OUUW01000005 SPP80589.1 AXCN02001157 CH902623 EDV30430.2 CM000070 EDY67822.2 JH432214 CH480828 EDW45216.1 CM000364 EDX14642.1 AJWK01024228 CH964232 EDW81120.2 CH954182 EDV53445.1 GANO01002751 JAB57120.1 KK855982 PTY24543.1 GECZ01006033 JAS63736.1 AJVK01029972 AJVK01029973 GFDF01009872 JAV04212.1 JXJN01015845 CCAG010023687 GALX01005899 JAB62567.1 CH479179 EDW24019.1 GEBQ01009301 JAT30676.1 GECZ01025413 JAS44356.1 GDIQ01240163 JAK11562.1 GDIQ01026604 JAN68133.1 LRGB01000781 KZS15957.1 KK852683 KDR18548.1 GFWV01022825 MAA47552.1 GEBQ01028285 JAT11692.1 GDIP01210699 JAJ12703.1 GDIP01204596 JAJ18806.1 GDIP01029050 JAM74665.1
KQ459596 BAM18570.1 KPI95267.1 GAIX01013951 JAA78609.1 ODYU01004030 SOQ43490.1 KQ460118 KPJ17706.1 KQ971338 EFA01723.1 UFQT01001990 SSX32322.1 UFQS01000630 UFQT01000630 SSX05547.1 SSX25906.1 GAMD01002789 JAA98801.1 SSX32321.1 SSX05546.1 SSX25905.1 GDHF01004037 JAI48277.1 GAKP01009703 GAKP01009702 GAKP01009701 GAKP01009700 JAC49249.1 GEHC01001290 JAV46355.1 GGFM01004945 MBW25696.1 GDAI01001055 JAI16548.1 GGFM01005008 MBW25759.1 GGFM01007724 MBW28475.1 GGFM01007797 MBW28548.1 ADMH02000944 ETN64603.1 GGFJ01008885 MBW58026.1 GFDG01001806 JAV16993.1 GFDG01001801 JAV16998.1 GGFJ01008883 MBW58024.1 CH477207 EAT47852.1 GFDG01001820 JAV16979.1 JXUM01021495 KQ560602 KXJ81678.1 KA645554 AFP60183.1 GGFK01008453 MBW41774.1 CH940650 EDW67271.1 APGK01026955 APGK01026956 KB740648 KB631780 ENN79735.1 ERL86037.1 JRES01001623 KNC21450.1 APCN01002552 GEZM01026068 GEZM01026067 JAV87264.1 CP012526 ALC46399.1 GAMC01009912 GAMC01009911 JAB96644.1 AAAB01008880 EAA08524.4 GFDL01007420 JAV27625.1 DS232039 EDS32803.1 AE014297 AAF56679.3 ALI30643.1 ATLV01026898 KE525420 KFB53550.1 CM000160 EDW98532.2 BT126160 ADZ99412.1 JXJN01007687 CH933806 KRG02070.1 CH916373 EDV94455.1 OUUW01000005 SPP80589.1 AXCN02001157 CH902623 EDV30430.2 CM000070 EDY67822.2 JH432214 CH480828 EDW45216.1 CM000364 EDX14642.1 AJWK01024228 CH964232 EDW81120.2 CH954182 EDV53445.1 GANO01002751 JAB57120.1 KK855982 PTY24543.1 GECZ01006033 JAS63736.1 AJVK01029972 AJVK01029973 GFDF01009872 JAV04212.1 JXJN01015845 CCAG010023687 GALX01005899 JAB62567.1 CH479179 EDW24019.1 GEBQ01009301 JAT30676.1 GECZ01025413 JAS44356.1 GDIQ01240163 JAK11562.1 GDIQ01026604 JAN68133.1 LRGB01000781 KZS15957.1 KK852683 KDR18548.1 GFWV01022825 MAA47552.1 GEBQ01028285 JAT11692.1 GDIP01210699 JAJ12703.1 GDIP01204596 JAJ18806.1 GDIP01029050 JAM74665.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000007266
+ More
UP000069272 UP000000673 UP000075881 UP000008820 UP000069940 UP000249989 UP000095301 UP000008792 UP000019118 UP000030742 UP000095300 UP000037069 UP000075884 UP000076407 UP000075840 UP000075880 UP000092553 UP000007062 UP000075885 UP000075902 UP000075903 UP000076408 UP000002320 UP000000803 UP000030765 UP000192221 UP000002282 UP000092445 UP000092443 UP000092460 UP000075920 UP000075900 UP000009192 UP000001070 UP000075882 UP000268350 UP000075886 UP000078200 UP000007801 UP000001819 UP000001292 UP000000304 UP000092461 UP000007798 UP000008711 UP000091820 UP000192223 UP000092462 UP000092444 UP000008744 UP000076858 UP000027135
UP000069272 UP000000673 UP000075881 UP000008820 UP000069940 UP000249989 UP000095301 UP000008792 UP000019118 UP000030742 UP000095300 UP000037069 UP000075884 UP000076407 UP000075840 UP000075880 UP000092553 UP000007062 UP000075885 UP000075902 UP000075903 UP000076408 UP000002320 UP000000803 UP000030765 UP000192221 UP000002282 UP000092445 UP000092443 UP000092460 UP000075920 UP000075900 UP000009192 UP000001070 UP000075882 UP000268350 UP000075886 UP000078200 UP000007801 UP000001819 UP000001292 UP000000304 UP000092461 UP000007798 UP000008711 UP000091820 UP000192223 UP000092462 UP000092444 UP000008744 UP000076858 UP000027135
Interpro
SUPFAM
SSF55811
SSF55811
ProteinModelPortal
H9J5I5
A0A3S2NSJ3
A0A212FBY6
I4DKY0
S4NNF5
A0A2H1VRP4
+ More
A0A194RNW1 D2A0B4 A0A336MPJ8 A0A336M6L8 T1DQ41 A0A336MRY2 A0A336KL19 A0A0K8WB32 A0A034W3H9 A0A1W7R5R8 A0A2M3ZAX5 A0A182FPU7 A0A0K8TQX5 A0A1S4EXV6 A0A2M3ZB57 A0A2M3ZJ31 A0A2M3ZJD1 W5JMR1 A0A2M4BY66 A0A1L8EEK4 A0A1L8EE70 A0A2M4BYD4 A0A182K1D8 Q17M97 A0A1L8EEF5 A0A182GYQ6 T1PA38 A0A2M4ALX7 B4M0A9 N6UDF7 A0A1I8Q3X0 A0A0L0BMX2 A0A182N5S7 A0A182XJ27 A0A182I7T2 A0A1Y1MNL6 A0A182JMI3 A0A0M3QXS9 W8C3Y4 Q7QB65 A0A182P1Q4 A0A1Q3FJC4 A0A182TRF0 A0A182V9H9 A0A182YGM5 B0WQD2 Q9VB64 A0A084WTK6 A0A1W4UIY6 B4PRT9 F2FB73 A0A240SX77 A0A1A9YSU7 A0A1B0B2P8 A0A182W1I8 A0A182RYM8 A0A0Q9X1U9 B4JRR5 A0A182L7C9 A0A3B0K3N3 A0A182QGF8 A0A1A9VFZ6 B3MT40 B5DWT8 T1JH04 B4IC58 B4QXF6 A0A1B0CQZ0 B4NBF6 B3P841 U5EWZ5 A0A1A9WF40 A0A1A9WR22 A0A1W4XS49 A0A2R7WYD3 A0A1B6GMR0 A0A1B0DBG7 A0A1L8DCN6 A0A1B0BK86 A0A1B0FCU4 V5GLU0 B4G2H4 A0A1B6M416 A0A1B6F258 A0A0N8AV12 A0A0N8EHD3 A0A164Z5B6 A0A067RHA2 A0A293N3C1 A0A1B6KJQ1 A0A0P4ZNX9 A0A0P5A5X9 A0A0P6AHW7
A0A194RNW1 D2A0B4 A0A336MPJ8 A0A336M6L8 T1DQ41 A0A336MRY2 A0A336KL19 A0A0K8WB32 A0A034W3H9 A0A1W7R5R8 A0A2M3ZAX5 A0A182FPU7 A0A0K8TQX5 A0A1S4EXV6 A0A2M3ZB57 A0A2M3ZJ31 A0A2M3ZJD1 W5JMR1 A0A2M4BY66 A0A1L8EEK4 A0A1L8EE70 A0A2M4BYD4 A0A182K1D8 Q17M97 A0A1L8EEF5 A0A182GYQ6 T1PA38 A0A2M4ALX7 B4M0A9 N6UDF7 A0A1I8Q3X0 A0A0L0BMX2 A0A182N5S7 A0A182XJ27 A0A182I7T2 A0A1Y1MNL6 A0A182JMI3 A0A0M3QXS9 W8C3Y4 Q7QB65 A0A182P1Q4 A0A1Q3FJC4 A0A182TRF0 A0A182V9H9 A0A182YGM5 B0WQD2 Q9VB64 A0A084WTK6 A0A1W4UIY6 B4PRT9 F2FB73 A0A240SX77 A0A1A9YSU7 A0A1B0B2P8 A0A182W1I8 A0A182RYM8 A0A0Q9X1U9 B4JRR5 A0A182L7C9 A0A3B0K3N3 A0A182QGF8 A0A1A9VFZ6 B3MT40 B5DWT8 T1JH04 B4IC58 B4QXF6 A0A1B0CQZ0 B4NBF6 B3P841 U5EWZ5 A0A1A9WF40 A0A1A9WR22 A0A1W4XS49 A0A2R7WYD3 A0A1B6GMR0 A0A1B0DBG7 A0A1L8DCN6 A0A1B0BK86 A0A1B0FCU4 V5GLU0 B4G2H4 A0A1B6M416 A0A1B6F258 A0A0N8AV12 A0A0N8EHD3 A0A164Z5B6 A0A067RHA2 A0A293N3C1 A0A1B6KJQ1 A0A0P4ZNX9 A0A0P5A5X9 A0A0P6AHW7
PDB
3Q91
E-value=1.88076e-29,
Score=319
Ontologies
GO
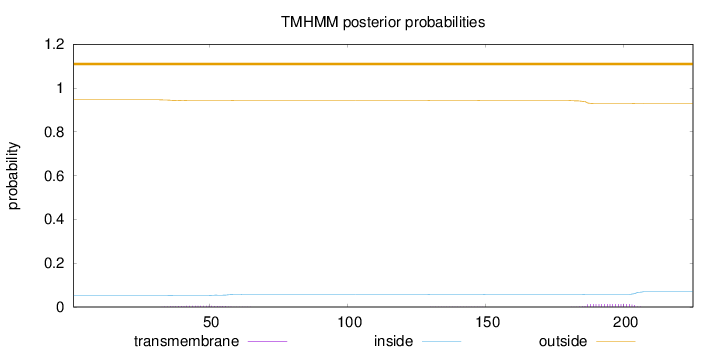
Topology
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35589
Exp number, first 60 AAs:
0.10246
Total prob of N-in:
0.05359
outside
1 - 225
Population Genetic Test Statistics
Pi
166.053911
Theta
180.709778
Tajima's D
-0.755425
CLR
1.16872
CSRT
0.188440577971101
Interpretation
Uncertain
