Pre Gene Modal
BGIBMGA004740
Annotation
DNA_damage-binding_protein_1_[Papilio_xuthus]
Full name
DNA damage-binding protein 1
Alternative Name
DDB p127 subunit
DNA damage-binding protein a
Damage-specific DNA-binding protein 1
HBV X-associated protein 1
UV-damaged DNA-binding factor
UV-damaged DNA-binding protein 1
XPE-binding factor
Xeroderma pigmentosum group E-complementing protein
DNA damage-binding protein a
Damage-specific DNA-binding protein 1
HBV X-associated protein 1
UV-damaged DNA-binding factor
UV-damaged DNA-binding protein 1
XPE-binding factor
Xeroderma pigmentosum group E-complementing protein
Location in the cell
Cytoplasmic Reliability : 2.341
Sequence
CDS
ATGGAAGTGTCGGTGGGGGGAGGGTTGGGGGGCGAGGTCGGTAACGGCTTGTGGGTCCACAGCGCGGCCACCACGGACGAGGACCGGCAGCAGATGGGCTACGCGGGGCAGTTCCACCTCGGCGACATGGTCAACGTCATGCGCGCCGGCTCCCTCGTGGCGCCGCACCACCACGCCGACTCCGCCGCGCCCACGCACCGCCCCGTGCTGCTCGCCACCGTGCACGGCGCCATCTTTTTAGTGGCTCAGATATCCCAGGAGCTGTTCGAGTTCCTGCAGCAGGTGGAGGAGCGGCTGTCGCACTGCATCAAGTCCGTGGGCCGCATCCCGCACGCCTTCTGGCGCTCCTTCAACACCGACATCAAGACCGAGCCCGCCGAGGGGTTCGTCGACGGGGACCTCATCGAGACCTTCCTCGACCTCAACAGGGACGTGCAGCACGACATCGTCAAGGGACTACAGATCGACGAGGGCAGCGGCATGATGAGGGACGCCAAAATAGACGACTTAATCAAAATAGTGGAAGACCTAACGAGGATACATTGA
Protein
MEVSVGGGLGGEVGNGLWVHSAATTDEDRQQMGYAGQFHLGDMVNVMRAGSLVAPHHHADSAAPTHRPVLLATVHGAIFLVAQISQELFEFLQQVEERLSHCIKSVGRIPHAFWRSFNTDIKTEPAEGFVDGDLIETFLDLNRDVQHDIVKGLQIDEGSGMMRDAKIDDLIKIVEDLTRIH
Summary
Description
Required for DNA repair. Binds to DDB2 to form the UV-damaged DNA-binding protein complex (the UV-DDB complex). The UV-DDB complex may recognize UV-induced DNA damage and recruit proteins of the nucleotide excision repair pathway (the NER pathway) to initiate DNA repair. The UV-DDB complex preferentially binds to cyclobutane pyrimidine dimers (CPD), 6-4 photoproducts (6-4 PP), apurinic sites and short mismatches. Also appears to function as a component of numerous distinct DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins. The functional specificity of the DCX E3 ubiquitin-protein ligase complex is determined by the variable substrate recognition component recruited by DDB1. DCX(DDB2) (also known as DDB1-CUL4-ROC1, CUL4-DDB-ROC1 and CUL4-DDB-RBX1) may ubiquitinate histone H2A, histone H3 and histone H4 at sites of UV-induced DNA damage. The ubiquitination of histones may facilitate their removal from the nucleosome and promote subsequent DNA repair. DCX(DDB2) also ubiquitinates XPC, which may enhance DNA-binding by XPC and promote NER. DCX(DTL) plays a role in PCNA-dependent polyubiquitination of CDT1 and MDM2-dependent ubiquitination of TP53 in response to radiation-induced DNA damage and during DNA replication. DCX(ERCC8) (the CSA complex) plays a role in transcription-coupled repair (TCR). May also play a role in ubiquitination of CDKN1B/p27kip when associated with CUL4 and SKP2. The DDB1-CUL4A-DTL E3 ligase complex regulates the circadian clock function by mediating the ubiquitination and degradation of CRY1 (PubMed:26431207). DDB1-mediated CRY1 degradation promotes FOXO1 protein stability and FOXO1-mediated gluconeogenesis in the liver (By similarity).
Subunit
Component of the UV-DDB complex which includes DDB1 and DDB2; the heterodimer dimerizes to give rise to a heterotetramer when bound to damaged DNA (PubMed:22822215). The UV-DDB complex interacts with monoubiquitinated histone H2A and binds to XPC via the DDB2 subunit. Component of numerous DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complexes which consist of a core of DDB1, CUL4A or CUL4B and RBX1. DDB1 may recruit specific substrate targeting subunits to the DCX complex. These substrate targeting subunits are generally known as DCAF (DDB1- and CUL4-associated factor) or CDW (CUL4-DDB1-associated WD40-repeat) proteins. Interacts with AMBRA1, ATG16L1, BTRC, CRBN, DCAF1, DCAF4, DCAF5, DCAF6, DCAF7, DCAF8, DCAF9, DCAF10, DCAF11, DCAF12, DCAF15, DCAF16, DCAF17, DDA1, DET1, DTL, ERCC8, FBXW5, FBXW8, GRWD1, KATNB1, NLE1, NUP43, PAFAH1B1, PHIP, PWP1, RBBP4, RBBP5, RBBP7, COP1, SNRNP40, DCAF1, WDR5, WDR5B, WDR12, WDR26, WDR39, WDR42, WDR53, WDR59, WDR61, WSB1, WSB2, LRWD1 and WDTC1. DCX complexes may associate with the COP9 signalosome, and this inhibits the E3 ubiquitin-protein ligase activity of the complex. Interacts with NF2, TSC1 and TSC2. Interaction with SV5 protein V may prevent the recruitment of DCAF proteins to DCX complexes. Interacts with AGO1 and AGO2. Associates with the E3 ligase complex containing DYRK2, EDD/UBR5, DDB1 and DCAF1 proteins (EDVP complex). Interacts directly with DYRK2. DCX(DTL) complex interacts with FBXO11; does not ubiquitinate and degradate FBXO11. Interacts with TRPC4AP (PubMed:19966799). Interacts with CRY1 and CRY2 (By similarity). The DDB1-CUL4A complex interacts with CRY1 (PubMed:26431207).
(Microbial infection) Interacts with Simian virus 5 protein V.
(Microbial infection) Interacts with hepatitis B virus protein HBX; the viral protein contains a short helical motif that competes for the same binding site as the N-terminal helical motif found in endogenous DCAF proteins.
(Microbial infection) Interacts with Simian virus 5 protein V.
(Microbial infection) Interacts with hepatitis B virus protein HBX; the viral protein contains a short helical motif that competes for the same binding site as the N-terminal helical motif found in endogenous DCAF proteins.
Similarity
Belongs to the DDB1 family.
Keywords
3D-structure
Acetylation
Alternative splicing
Biological rhythms
Complete proteome
Cytoplasm
DNA damage
DNA repair
DNA-binding
Host-virus interaction
Isopeptide bond
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Repeat
Ubl conjugation
Ubl conjugation pathway
Feature
chain DNA damage-binding protein 1
splice variant In isoform 2.
sequence variant In dbSNP:rs28720299.
splice variant In isoform 2.
sequence variant In dbSNP:rs28720299.
Uniprot
H9J5F0
A0A2H1WKE8
S4NWY3
A0A194PP99
A0A194RJH7
A0A212FBZ7
+ More
A0A2A4JZ82 A0A3S2LJK0 A0A2J7R6I6 A0A2Z5TRE2 A0A2J7R6I4 A0A067QWW9 A0A1B6IJV9 A0A1B6I554 A0A1B6ETQ7 A0A2P8YXD9 L7LV98 L7M756 A0A1B6DCC1 A0A131XUS2 A0A210PZE9 V9IJQ1 K1RKZ5 A0A232F584 A0A224YX42 K7IUN5 A0A131YKZ0 A0A131XG70 A0A1E1XLY5 A0A0C9RQP9 U5EVD6 A0A2D4KBE6 A0A0P5HHR3 A0A0P5RE31 A0A1E1XAG8 A0A0P6HGV3 A0A0P6EYQ9 E0VJ24 A0A0P5RR17 A0A0P6FCL2 A0A0P5RXC9 A0A0P6FRV4 A0A0P5R5T6 A0A0P5LWX0 A0A164LX96 E9FU30 R7UIH1 A0A151JS91 A0A195EXY0 A0A158NJ58 A0A293MTW8 A0A0P6GZA2 A0A0P5LCR1 A0A1Z5LES1 A0A195C340 A0A151X0H4 F4W766 A0A195BL46 E2BJY1 A0A0P6C9R4 A0A0P5IFL8 A0A0P5AAI7 A0A2R5L5Y4 A0A154PI86 A0A0P5KLL0 B7PIF3 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A026WBL0 A0A1S3IIX8 A0A1S3IIM7 E2A411 A0A0J7L6L1 A0A2R7W2L8 C3ZMJ6 A8E561 U6DWG1 A0A1W4X581 A0A087SY87 V9K9R8 Q16531-2 A0A0P6GFZ4 K7G405 F7FU40 A0A0L7R1I9 C0PUD7 A0A151NYZ5 A0A0B7AQY1 A0A0F7Z6E4 A0A0B8RR16 A0A098LZ59 A0A0F7ZE80 D6WR91 A0A3Q0GSF5 A0A1W7RHX9 A0A146LUG5 T1DLK0 Q3ULS8 H9GPM9
A0A2A4JZ82 A0A3S2LJK0 A0A2J7R6I6 A0A2Z5TRE2 A0A2J7R6I4 A0A067QWW9 A0A1B6IJV9 A0A1B6I554 A0A1B6ETQ7 A0A2P8YXD9 L7LV98 L7M756 A0A1B6DCC1 A0A131XUS2 A0A210PZE9 V9IJQ1 K1RKZ5 A0A232F584 A0A224YX42 K7IUN5 A0A131YKZ0 A0A131XG70 A0A1E1XLY5 A0A0C9RQP9 U5EVD6 A0A2D4KBE6 A0A0P5HHR3 A0A0P5RE31 A0A1E1XAG8 A0A0P6HGV3 A0A0P6EYQ9 E0VJ24 A0A0P5RR17 A0A0P6FCL2 A0A0P5RXC9 A0A0P6FRV4 A0A0P5R5T6 A0A0P5LWX0 A0A164LX96 E9FU30 R7UIH1 A0A151JS91 A0A195EXY0 A0A158NJ58 A0A293MTW8 A0A0P6GZA2 A0A0P5LCR1 A0A1Z5LES1 A0A195C340 A0A151X0H4 F4W766 A0A195BL46 E2BJY1 A0A0P6C9R4 A0A0P5IFL8 A0A0P5AAI7 A0A2R5L5Y4 A0A154PI86 A0A0P5KLL0 B7PIF3 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A026WBL0 A0A1S3IIX8 A0A1S3IIM7 E2A411 A0A0J7L6L1 A0A2R7W2L8 C3ZMJ6 A8E561 U6DWG1 A0A1W4X581 A0A087SY87 V9K9R8 Q16531-2 A0A0P6GFZ4 K7G405 F7FU40 A0A0L7R1I9 C0PUD7 A0A151NYZ5 A0A0B7AQY1 A0A0F7Z6E4 A0A0B8RR16 A0A098LZ59 A0A0F7ZE80 D6WR91 A0A3Q0GSF5 A0A1W7RHX9 A0A146LUG5 T1DLK0 Q3ULS8 H9GPM9
Pubmed
19121390
23622113
26354079
22118469
26760975
24845553
+ More
29403074 25576852 28812685 22992520 28648823 28797301 20075255 26830274 28049606 29209593 28503490 20566863 21292972 23254933 21347285 28528879 21719571 20798317 24508170 30249741 18563158 12477932 24402279 8530102 7815490 8538642 14702039 16554811 15489334 9632823 10777491 11673459 11531405 12732143 12743284 15448697 14739464 15882621 16223728 16227264 16260596 16482215 17079684 16407242 16407252 16527807 16678110 16949367 16940174 17041588 16473935 17932509 18593899 18381890 18606781 18332868 19413330 19287380 19608861 21269460 22935713 23478445 24275569 26431207 28790135 28112733 16413485 16964240 19109893 19966799 22118460 21468892 22822215 25043012 25108355 17381049 18464734 22293439 25476704 18362917 19820115 26358130 26823975 23758969 25727380 10349636 11042159 11076861 11217851 12466851 16141073 21881562
29403074 25576852 28812685 22992520 28648823 28797301 20075255 26830274 28049606 29209593 28503490 20566863 21292972 23254933 21347285 28528879 21719571 20798317 24508170 30249741 18563158 12477932 24402279 8530102 7815490 8538642 14702039 16554811 15489334 9632823 10777491 11673459 11531405 12732143 12743284 15448697 14739464 15882621 16223728 16227264 16260596 16482215 17079684 16407242 16407252 16527807 16678110 16949367 16940174 17041588 16473935 17932509 18593899 18381890 18606781 18332868 19413330 19287380 19608861 21269460 22935713 23478445 24275569 26431207 28790135 28112733 16413485 16964240 19109893 19966799 22118460 21468892 22822215 25043012 25108355 17381049 18464734 22293439 25476704 18362917 19820115 26358130 26823975 23758969 25727380 10349636 11042159 11076861 11217851 12466851 16141073 21881562
EMBL
BABH01028209
BABH01028210
BABH01028211
BABH01028212
ODYU01009154
SOQ53382.1
+ More
GAIX01012372 JAA80188.1 KQ459596 KPI95266.1 KQ460118 KPJ17707.1 AGBW02009250 OWR51255.1 NWSH01000383 PCG76813.1 RSAL01000001 RVE55275.1 NEVH01006756 PNF36444.1 FX985782 BBA93669.1 PNF36443.1 KK853153 KDR10581.1 GECU01020496 JAS87210.1 GECU01025672 JAS82034.1 GECZ01028511 JAS41258.1 PYGN01000302 PSN48913.1 GACK01009507 JAA55527.1 GACK01005342 JAA59692.1 GEDC01013957 JAS23341.1 GEFM01005389 JAP70407.1 NEDP02005356 OWF41789.1 JR049167 AEY60882.1 JH817518 EKC34976.1 NNAY01000904 OXU25996.1 GFPF01007246 MAA18392.1 AAZX01002644 GEDV01009976 JAP78581.1 GEFH01003870 JAP64711.1 GFAA01003094 JAU00341.1 GBYB01010825 JAG80592.1 GANO01001076 JAB58795.1 IACL01047310 LAB06002.1 GDIQ01227878 JAK23847.1 GDIQ01102720 JAL49006.1 GFAC01003127 JAT96061.1 GDIQ01019023 JAN75714.1 GDIQ01056143 JAN38594.1 DS235219 EEB13380.1 GDIQ01106321 JAL45405.1 GDIQ01058842 JAN35895.1 GDIQ01095023 JAL56703.1 GDIQ01145109 GDIQ01061387 GDIQ01034995 JAN33350.1 GDIQ01112270 JAL39456.1 GDIQ01174390 JAK77335.1 LRGB01003123 KZS04522.1 GL732524 EFX89472.1 AMQN01008593 AMQN01008594 KB303505 ELU03057.1 KQ978579 KYN30011.1 KQ981920 KYN33006.1 ADTU01017499 GFWV01019585 MAA44313.1 GDIQ01038902 JAN55835.1 GDIQ01171396 JAK80329.1 GFJQ02001064 JAW05906.1 KQ978317 KYM95279.1 KQ982612 KYQ53861.1 GL887813 EGI69950.1 KQ976453 KYM85435.1 GL448708 EFN83990.1 GDIP01003641 JAN00075.1 GDIQ01214033 JAK37692.1 GDIP01202767 GDIP01030642 JAJ20635.1 GGLE01000766 MBY04892.1 KQ434905 KZC11204.1 GDIQ01182249 JAK69476.1 ABJB010287562 ABJB010324138 ABJB010534196 ABJB010642340 ABJB010687854 ABJB010731674 ABJB010884463 ABJB010898466 ABJB010908998 ABJB011009818 DS718559 EEC06375.1 KQ435863 KOX70388.1 KZ288194 PBC33915.1 KK107293 QOIP01000005 EZA53445.1 RLU22890.1 GL436519 EFN71863.1 LBMM01000453 KMQ98301.1 KK854164 PTY12415.1 GG666646 EEN46206.1 BC153473 AAI53474.1 HAAF01014436 CCP86258.1 KK112505 KFM57826.1 JW862087 AFO94604.1 U18299 L40326 U32986 AJ002955 AK294341 AK312436 AY960579 AP003037 AP003108 CH471076 BC011686 BC050530 BC051764 GDIQ01044145 JAN50592.1 AGCU01001011 AGCU01001012 AGCU01001013 KQ414667 KOC64712.1 BT072289 ACN58665.1 AKHW03001628 KYO41665.1 HACG01036107 CEK82972.1 GBEW01000291 JAI10074.1 GBSH01000871 JAG68154.1 GBSI01000729 JAC95767.1 GBEX01000981 JAI13579.1 KQ971354 EFA07036.1 GDAY02000691 JAV50736.1 GDHC01007884 JAQ10745.1 GAAZ01000708 GBKC01001015 GBKD01000636 JAA97235.1 JAG45055.1 JAG46982.1 AK145328 BAE26370.1 AAWZ02021149
GAIX01012372 JAA80188.1 KQ459596 KPI95266.1 KQ460118 KPJ17707.1 AGBW02009250 OWR51255.1 NWSH01000383 PCG76813.1 RSAL01000001 RVE55275.1 NEVH01006756 PNF36444.1 FX985782 BBA93669.1 PNF36443.1 KK853153 KDR10581.1 GECU01020496 JAS87210.1 GECU01025672 JAS82034.1 GECZ01028511 JAS41258.1 PYGN01000302 PSN48913.1 GACK01009507 JAA55527.1 GACK01005342 JAA59692.1 GEDC01013957 JAS23341.1 GEFM01005389 JAP70407.1 NEDP02005356 OWF41789.1 JR049167 AEY60882.1 JH817518 EKC34976.1 NNAY01000904 OXU25996.1 GFPF01007246 MAA18392.1 AAZX01002644 GEDV01009976 JAP78581.1 GEFH01003870 JAP64711.1 GFAA01003094 JAU00341.1 GBYB01010825 JAG80592.1 GANO01001076 JAB58795.1 IACL01047310 LAB06002.1 GDIQ01227878 JAK23847.1 GDIQ01102720 JAL49006.1 GFAC01003127 JAT96061.1 GDIQ01019023 JAN75714.1 GDIQ01056143 JAN38594.1 DS235219 EEB13380.1 GDIQ01106321 JAL45405.1 GDIQ01058842 JAN35895.1 GDIQ01095023 JAL56703.1 GDIQ01145109 GDIQ01061387 GDIQ01034995 JAN33350.1 GDIQ01112270 JAL39456.1 GDIQ01174390 JAK77335.1 LRGB01003123 KZS04522.1 GL732524 EFX89472.1 AMQN01008593 AMQN01008594 KB303505 ELU03057.1 KQ978579 KYN30011.1 KQ981920 KYN33006.1 ADTU01017499 GFWV01019585 MAA44313.1 GDIQ01038902 JAN55835.1 GDIQ01171396 JAK80329.1 GFJQ02001064 JAW05906.1 KQ978317 KYM95279.1 KQ982612 KYQ53861.1 GL887813 EGI69950.1 KQ976453 KYM85435.1 GL448708 EFN83990.1 GDIP01003641 JAN00075.1 GDIQ01214033 JAK37692.1 GDIP01202767 GDIP01030642 JAJ20635.1 GGLE01000766 MBY04892.1 KQ434905 KZC11204.1 GDIQ01182249 JAK69476.1 ABJB010287562 ABJB010324138 ABJB010534196 ABJB010642340 ABJB010687854 ABJB010731674 ABJB010884463 ABJB010898466 ABJB010908998 ABJB011009818 DS718559 EEC06375.1 KQ435863 KOX70388.1 KZ288194 PBC33915.1 KK107293 QOIP01000005 EZA53445.1 RLU22890.1 GL436519 EFN71863.1 LBMM01000453 KMQ98301.1 KK854164 PTY12415.1 GG666646 EEN46206.1 BC153473 AAI53474.1 HAAF01014436 CCP86258.1 KK112505 KFM57826.1 JW862087 AFO94604.1 U18299 L40326 U32986 AJ002955 AK294341 AK312436 AY960579 AP003037 AP003108 CH471076 BC011686 BC050530 BC051764 GDIQ01044145 JAN50592.1 AGCU01001011 AGCU01001012 AGCU01001013 KQ414667 KOC64712.1 BT072289 ACN58665.1 AKHW03001628 KYO41665.1 HACG01036107 CEK82972.1 GBEW01000291 JAI10074.1 GBSH01000871 JAG68154.1 GBSI01000729 JAC95767.1 GBEX01000981 JAI13579.1 KQ971354 EFA07036.1 GDAY02000691 JAV50736.1 GDHC01007884 JAQ10745.1 GAAZ01000708 GBKC01001015 GBKD01000636 JAA97235.1 JAG45055.1 JAG46982.1 AK145328 BAE26370.1 AAWZ02021149
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000283053
+ More
UP000235965 UP000027135 UP000245037 UP000242188 UP000005408 UP000215335 UP000002358 UP000009046 UP000076858 UP000000305 UP000014760 UP000078492 UP000078541 UP000005205 UP000078542 UP000075809 UP000007755 UP000078540 UP000008237 UP000076502 UP000001555 UP000053105 UP000242457 UP000005203 UP000053097 UP000279307 UP000085678 UP000000311 UP000036403 UP000001554 UP000192223 UP000054359 UP000005640 UP000007267 UP000002279 UP000053825 UP000050525 UP000007266 UP000189705 UP000001646
UP000235965 UP000027135 UP000245037 UP000242188 UP000005408 UP000215335 UP000002358 UP000009046 UP000076858 UP000000305 UP000014760 UP000078492 UP000078541 UP000005205 UP000078542 UP000075809 UP000007755 UP000078540 UP000008237 UP000076502 UP000001555 UP000053105 UP000242457 UP000005203 UP000053097 UP000279307 UP000085678 UP000000311 UP000036403 UP000001554 UP000192223 UP000054359 UP000005640 UP000007267 UP000002279 UP000053825 UP000050525 UP000007266 UP000189705 UP000001646
Interpro
Gene 3D
ProteinModelPortal
H9J5F0
A0A2H1WKE8
S4NWY3
A0A194PP99
A0A194RJH7
A0A212FBZ7
+ More
A0A2A4JZ82 A0A3S2LJK0 A0A2J7R6I6 A0A2Z5TRE2 A0A2J7R6I4 A0A067QWW9 A0A1B6IJV9 A0A1B6I554 A0A1B6ETQ7 A0A2P8YXD9 L7LV98 L7M756 A0A1B6DCC1 A0A131XUS2 A0A210PZE9 V9IJQ1 K1RKZ5 A0A232F584 A0A224YX42 K7IUN5 A0A131YKZ0 A0A131XG70 A0A1E1XLY5 A0A0C9RQP9 U5EVD6 A0A2D4KBE6 A0A0P5HHR3 A0A0P5RE31 A0A1E1XAG8 A0A0P6HGV3 A0A0P6EYQ9 E0VJ24 A0A0P5RR17 A0A0P6FCL2 A0A0P5RXC9 A0A0P6FRV4 A0A0P5R5T6 A0A0P5LWX0 A0A164LX96 E9FU30 R7UIH1 A0A151JS91 A0A195EXY0 A0A158NJ58 A0A293MTW8 A0A0P6GZA2 A0A0P5LCR1 A0A1Z5LES1 A0A195C340 A0A151X0H4 F4W766 A0A195BL46 E2BJY1 A0A0P6C9R4 A0A0P5IFL8 A0A0P5AAI7 A0A2R5L5Y4 A0A154PI86 A0A0P5KLL0 B7PIF3 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A026WBL0 A0A1S3IIX8 A0A1S3IIM7 E2A411 A0A0J7L6L1 A0A2R7W2L8 C3ZMJ6 A8E561 U6DWG1 A0A1W4X581 A0A087SY87 V9K9R8 Q16531-2 A0A0P6GFZ4 K7G405 F7FU40 A0A0L7R1I9 C0PUD7 A0A151NYZ5 A0A0B7AQY1 A0A0F7Z6E4 A0A0B8RR16 A0A098LZ59 A0A0F7ZE80 D6WR91 A0A3Q0GSF5 A0A1W7RHX9 A0A146LUG5 T1DLK0 Q3ULS8 H9GPM9
A0A2A4JZ82 A0A3S2LJK0 A0A2J7R6I6 A0A2Z5TRE2 A0A2J7R6I4 A0A067QWW9 A0A1B6IJV9 A0A1B6I554 A0A1B6ETQ7 A0A2P8YXD9 L7LV98 L7M756 A0A1B6DCC1 A0A131XUS2 A0A210PZE9 V9IJQ1 K1RKZ5 A0A232F584 A0A224YX42 K7IUN5 A0A131YKZ0 A0A131XG70 A0A1E1XLY5 A0A0C9RQP9 U5EVD6 A0A2D4KBE6 A0A0P5HHR3 A0A0P5RE31 A0A1E1XAG8 A0A0P6HGV3 A0A0P6EYQ9 E0VJ24 A0A0P5RR17 A0A0P6FCL2 A0A0P5RXC9 A0A0P6FRV4 A0A0P5R5T6 A0A0P5LWX0 A0A164LX96 E9FU30 R7UIH1 A0A151JS91 A0A195EXY0 A0A158NJ58 A0A293MTW8 A0A0P6GZA2 A0A0P5LCR1 A0A1Z5LES1 A0A195C340 A0A151X0H4 F4W766 A0A195BL46 E2BJY1 A0A0P6C9R4 A0A0P5IFL8 A0A0P5AAI7 A0A2R5L5Y4 A0A154PI86 A0A0P5KLL0 B7PIF3 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A026WBL0 A0A1S3IIX8 A0A1S3IIM7 E2A411 A0A0J7L6L1 A0A2R7W2L8 C3ZMJ6 A8E561 U6DWG1 A0A1W4X581 A0A087SY87 V9K9R8 Q16531-2 A0A0P6GFZ4 K7G405 F7FU40 A0A0L7R1I9 C0PUD7 A0A151NYZ5 A0A0B7AQY1 A0A0F7Z6E4 A0A0B8RR16 A0A098LZ59 A0A0F7ZE80 D6WR91 A0A3Q0GSF5 A0A1W7RHX9 A0A146LUG5 T1DLK0 Q3ULS8 H9GPM9
PDB
6H0G
E-value=6.14228e-43,
Score=434
Ontologies
PATHWAY
GO
GO:0005634
GO:0006281
GO:0003676
GO:0005524
GO:0004672
GO:0003684
GO:0043161
GO:0010564
GO:0043066
GO:0005615
GO:0070062
GO:0044877
GO:0051702
GO:0030674
GO:0045732
GO:0006295
GO:0046726
GO:0097602
GO:0010498
GO:1901990
GO:0042769
GO:0006974
GO:0006296
GO:0048511
GO:0016032
GO:0045070
GO:0006511
GO:0045722
GO:0000784
GO:0070911
GO:0000717
GO:0006289
GO:0031465
GO:0042752
GO:0016055
GO:0006283
GO:0003677
GO:0033683
GO:0016567
GO:0032991
GO:0031464
GO:0035518
GO:0000715
GO:0005737
GO:0005654
GO:0070914
GO:0043687
GO:0080008
GO:1902188
GO:0006294
GO:0071987
GO:0006293
GO:0003700
GO:0008270
GO:0043565
GO:0003707
GO:0006259
GO:0005515
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
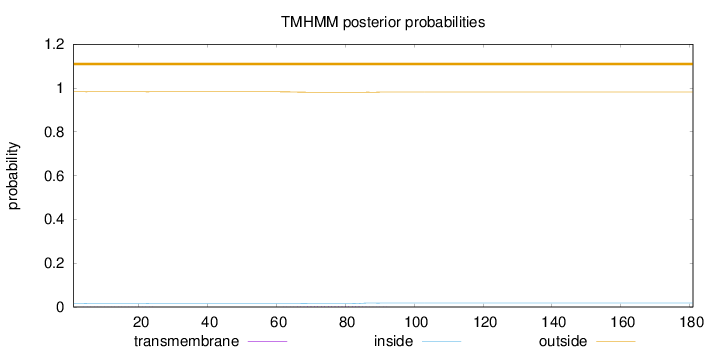
Length:
181
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08141
Exp number, first 60 AAs:
0.01369
Total prob of N-in:
0.01689
outside
1 - 181
Population Genetic Test Statistics
Pi
184.614469
Theta
177.968568
Tajima's D
-1.01961
CLR
0.163248
CSRT
0.126593670316484
Interpretation
Uncertain
