Gene
KWMTBOMO15422 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004740
Annotation
PREDICTED:_DNA_damage-binding_protein_1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.837 PlasmaMembrane Reliability : 1.744
Sequence
CDS
ATGTCTTACCACTACGTGGTCACAGCCCAAAAACAGACTTCGGTCACAGCATGTGTAACAGGAAACTTCACATCACCAACAGACCTAAATCTACTAGTAGCAAAGGTGTCTCGCCTGGAGATGTACTTGGTCACACCAGAAGGACTTCGCCCCATGAAGGAAGTTGGTTTATATGGAAAAGTTGCAAAGATGAAACTCTTTAGACCACCGTATGAAAATAAAGATCTAGTATTTATTCTAACTGCAAGATACAATGCCATGATATTAGAATGGCGTACAGGTGCAAATGGTGATCTCGAAGTTGTGACACGTGCACATGGTAATGTATCCGATCGAATTGGGAAACCATCAGAGAACGGCATCCTAGTTGTGATAGATCCCCAGGCAAGGGTCATCGGGTTGAGGCTGTATGACGGTCTCTTCAAGATTATACCATTGGAGAAGGACTCAGCAGAGCTGAAGGCTGCCAGCTTGAGGTTAGAAGAATTGAATGTTTACGATGTCGAATTCCTTCATGGATGTGCGAATCCGACATTGATACTGATCCATCAGGATACAAATGGGAGACACATTAAGACACGCGAGATAGGATTAAGAGATAAAGACTTTATAAAAGTATCATGGAAACAGGATAATGTTGAAACTGAAGCATCCATACTCATTCCGGTTCCGAGTCCACTAGGCGGAGCCATAGTTATTGGTCAAGAATCTATAGTGTATCACGACGGTCAGAGCTATGTGGCTGTCGCTCCACCGCAGATCAAGATGACACCTATCAACTGTTACTGCCGCGTGGACAGCCGCGGCCTGCGCTACCTAGCCGGCGACATCGCGGGGAGACTGTTCATGCTGCTGCTGGAGTTACACGAGAAGATGGACGGCACGCAGGCCGTCAAGGACCTCAAGGTCGAGCTCTTAGGCGACATCCCCATCCCGGAGTGCATGACGTACCTGGACAACGGGGTCGTGTTCATCGGCTCCCGGCTGGGCGACTCCGCGCTGGTCCGCCTGTCCAGCTCCCGGGACGACACTCACCAGTACGTGCTGCCCATGGAGACGTTCACCAGCCTCGCGCCCATCGTCGACATGTGCGTCGTGGACCTCGAGCGCCAAGGACAGAGCCAGCTCATCACCTGCTCCGGGGCGTTCAAAATGGGTTCGTTACGGATCATCCGCAACGGGATCGGCATCCAGGAGCAGGCCTCCATCGACCTGCCCGGCATCAAGGGCATGTGGCCCCTCACCCTCGCCCCGGACTCCGCCCACCACGACACCCTGGTGCTGTCCTTTGTCGGACAAACTAGGGTGCTGGCGCTTAATGGCGAAGAAGTCGAAGAGACGGAGATCAAGGGGTTCCTCTCGGACAGACAGACGTTCTTCACAGGCAACGTATGTTACGACCAGCTGATACAGGTGACGGACGAGGGCGTGCGGCTGGCGGGTCGCGCGGGCGCGGGGTGGTGCGTGACGTCACAGTGGCGCGCGCCCGCCGCGCTGTCCGTGGTGGCGTGCGCGCGCAGCCGCCTCGTGTGCGCCGCCGGCCCGCGCCTCTACCTGCTGCTGCTGCACGAAGCCGCCATCGAGCTCAAGGCCGAGGTGTGCATGGAGGAGGAGGTGGCGTGCCTGGACCTGGGCCCCGGCGAGGAGCAGGCGCTGCTGGGCGTGGGGCTGTGGACCGACATCTCCGTGCGCGTGCTGCGCCTGCCCGACATGACGCCGCTGCACACCGAGAAGCTGTACGGAGAGATAATACCGCGGTCGCTCATCATCAGCGTGCTGGAGGGCGTCTGCTACCTGCTGTGCGCGCTGGGGGACGGCTCCATGTTCTACTTCACCGTCGACCAGAAGACCGGAGTGCTGTCCAACAAGAAGAAGGTGACCCTCGGCACCCAGCCCACCGTGCTCAGGACCTTCAGGTCGCTGTCCACGACGAACATATTCGCGTGCTCGGACCGGCCCACCGTGATATTCTCGTCCAACCACAAGCTCGTGTTCTCCAACGTCAACCTCAAGGAGGTCACGCACATGTGCTCGCTCAACGCGCAGGCCTACCCCGACAGCCTGGCGCTGGCCACGGACAGCACGGTGACCATCGGCACCATCGACGAGATCCAGAAGCTGCACATCCGCACCGTGCCGCTGGGGGAGACGCCGCGCAGGATCGCCTACCAGGAGGCCTCGCAGACCTTCGCCGTGATAACGATGCGCGTGGAGAAGCTGGAGTGGGGCGGCGGCGAGGCGGGGCGGGCGCTGGACCACGAGCTGCACAACCTGCTCATACTCGACCACCACACCTTCGAAGTGCTGCACTCGCACCAGCTGATGTCGAGCGAGCTGGCCATGTCGCTGATGTCGTGCCGCCTGGGGGACGACCCCGCGCACTACTACGCCGTGGGGACCGCCATACTCAACCCTGAGGAGCCGGAGCCTAAAAAGGGCAGGATACTGCTGTTCCACTGGTCCGAAGGGAAACTGGTACAAGTAGCCGAAAAGGAAATCAAAGGGGGCTGCTACTCGCTGGTGGAGTTCAACGGCAAGCTACTGGCCGCCATCAACAGCACGGTGCGGCTGTTTGAGTGGACGTCGGACAAGGAGCTGCGGCTGGAGTGCAGTCACTTCAACAACATCGTGGCTCTGTACCTGAAGGCGAAGGGAGACTTCATCCTGGTCGGGGATCTGATGCGCTCCATGTCGCTACTGCAATATAAACAGATGGAGGGCAGCTTCGAGGAGATAGCCCGCGACTACAGCCCCAACTGGATGACGGCCGTGGAGATCCTGGACGACGACACGTTCCTGGGCGCAGAGAACAGCTTCAACCTTTTCGTTTGTCAAAAGGACAGGTGA
Protein
MSYHYVVTAQKQTSVTACVTGNFTSPTDLNLLVAKVSRLEMYLVTPEGLRPMKEVGLYGKVAKMKLFRPPYENKDLVFILTARYNAMILEWRTGANGDLEVVTRAHGNVSDRIGKPSENGILVVIDPQARVIGLRLYDGLFKIIPLEKDSAELKAASLRLEELNVYDVEFLHGCANPTLILIHQDTNGRHIKTREIGLRDKDFIKVSWKQDNVETEASILIPVPSPLGGAIVIGQESIVYHDGQSYVAVAPPQIKMTPINCYCRVDSRGLRYLAGDIAGRLFMLLLELHEKMDGTQAVKDLKVELLGDIPIPECMTYLDNGVVFIGSRLGDSALVRLSSSRDDTHQYVLPMETFTSLAPIVDMCVVDLERQGQSQLITCSGAFKMGSLRIIRNGIGIQEQASIDLPGIKGMWPLTLAPDSAHHDTLVLSFVGQTRVLALNGEEVEETEIKGFLSDRQTFFTGNVCYDQLIQVTDEGVRLAGRAGAGWCVTSQWRAPAALSVVACARSRLVCAAGPRLYLLLLHEAAIELKAEVCMEEEVACLDLGPGEEQALLGVGLWTDISVRVLRLPDMTPLHTEKLYGEIIPRSLIISVLEGVCYLLCALGDGSMFYFTVDQKTGVLSNKKKVTLGTQPTVLRTFRSLSTTNIFACSDRPTVIFSSNHKLVFSNVNLKEVTHMCSLNAQAYPDSLALATDSTVTIGTIDEIQKLHIRTVPLGETPRRIAYQEASQTFAVITMRVEKLEWGGGEAGRALDHELHNLLILDHHTFEVLHSHQLMSSELAMSLMSCRLGDDPAHYYAVGTAILNPEEPEPKKGRILLFHWSEGKLVQVAEKEIKGGCYSLVEFNGKLLAAINSTVRLFEWTSDKELRLECSHFNNIVALYLKAKGDFILVGDLMRSMSLLQYKQMEGSFEEIARDYSPNWMTAVEILDDDTFLGAENSFNLFVCQKDR
Summary
Uniprot
A0A2A4JZ82
A0A212FBZ7
A0A3S2LJK0
A0A194PP99
A0A194RJH7
A0A2Z5TRE2
+ More
A0A2J7R6I4 A0A067QWW9 A0A1L8DGY1 E9J1W0 H9J5F0 A0A158NJ58 A0A195BL46 A0A195EXY0 A0A151X0H4 F4W766 A0A195C340 A0A026WBL0 A0A151JS91 E2BJY1 A0A0C9RQP9 A0A0L7R1I9 E2A411 A0A232F584 A0A1Y1K0V9 K7IUN5 A0A1B0CUN8 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A154PI86 A0A1W4X581 A0A0J7L6L1 S4PYF2 D6WR91 Q17I86 E0VJ24 Q17I87 U5EVD6 A0A1S4F1P1 A0A336LNQ7 A0A182H165 A0A2M4A7Y9 W5JL18 A0A1J1I7F1 A0A182FT49 A0A1Q3F644 B0X4E5 A0A084WRC6 A0A182XWW1 A0A182NLA8 A0A2H1WKD0 A0A182UN77 A0A182UGX6 A0A182LF28 A0A182XI06 Q7QC66 A0A182HSY7 A0A182VXE5 A0A182JRI8 A0A182MI35 A0A182QID4 A0A182PAF4 A0A1S3IIM7 A0A2M4BAV2 N6UAL4 A0A182JC38 A0A1S3IIX8 E9FU30 A0A182R366 A0A0P5KLL0 A0A0P6GFZ4 A0A0P6FRV4 A0A0P5IFL8 A0A0P5RR17 A0A0P6E5N7 A0A0P5AAI7 V4C8B8 A0A0N7ZL83 A0A0P5LWX0 A0A0P5RXC9 A0A0P6FCL2 C3ZMJ6 A0A060X6D7 A0A2K6L728 A0A2J8LD23 A0A2J8SMZ1 F7HN48 B7Z2A1 F5GY55 C0H989 A0A3P8Z8J6 F6QDR0 V9K9R8 A0A286XPJ3 A0A2K6BUZ0 A0A2K6L717 L5KQI8 A0A2K5VZB4 G3QN74
A0A2J7R6I4 A0A067QWW9 A0A1L8DGY1 E9J1W0 H9J5F0 A0A158NJ58 A0A195BL46 A0A195EXY0 A0A151X0H4 F4W766 A0A195C340 A0A026WBL0 A0A151JS91 E2BJY1 A0A0C9RQP9 A0A0L7R1I9 E2A411 A0A232F584 A0A1Y1K0V9 K7IUN5 A0A1B0CUN8 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A154PI86 A0A1W4X581 A0A0J7L6L1 S4PYF2 D6WR91 Q17I86 E0VJ24 Q17I87 U5EVD6 A0A1S4F1P1 A0A336LNQ7 A0A182H165 A0A2M4A7Y9 W5JL18 A0A1J1I7F1 A0A182FT49 A0A1Q3F644 B0X4E5 A0A084WRC6 A0A182XWW1 A0A182NLA8 A0A2H1WKD0 A0A182UN77 A0A182UGX6 A0A182LF28 A0A182XI06 Q7QC66 A0A182HSY7 A0A182VXE5 A0A182JRI8 A0A182MI35 A0A182QID4 A0A182PAF4 A0A1S3IIM7 A0A2M4BAV2 N6UAL4 A0A182JC38 A0A1S3IIX8 E9FU30 A0A182R366 A0A0P5KLL0 A0A0P6GFZ4 A0A0P6FRV4 A0A0P5IFL8 A0A0P5RR17 A0A0P6E5N7 A0A0P5AAI7 V4C8B8 A0A0N7ZL83 A0A0P5LWX0 A0A0P5RXC9 A0A0P6FCL2 C3ZMJ6 A0A060X6D7 A0A2K6L728 A0A2J8LD23 A0A2J8SMZ1 F7HN48 B7Z2A1 F5GY55 C0H989 A0A3P8Z8J6 F6QDR0 V9K9R8 A0A286XPJ3 A0A2K6BUZ0 A0A2K6L717 L5KQI8 A0A2K5VZB4 G3QN74
Pubmed
22118469
26354079
26760975
24845553
21282665
19121390
+ More
21347285 21719571 24508170 30249741 20798317 28648823 28004739 20075255 23622113 18362917 19820115 17510324 20566863 26483478 20920257 23761445 24438588 25244985 20966253 12364791 14747013 17210077 23537049 21292972 23254933 18563158 24755649 17431167 16554811 19413330 21269460 24275569 20433749 25069045 19892987 24402279 21993624 23258410 22398555
21347285 21719571 24508170 30249741 20798317 28648823 28004739 20075255 23622113 18362917 19820115 17510324 20566863 26483478 20920257 23761445 24438588 25244985 20966253 12364791 14747013 17210077 23537049 21292972 23254933 18563158 24755649 17431167 16554811 19413330 21269460 24275569 20433749 25069045 19892987 24402279 21993624 23258410 22398555
EMBL
NWSH01000383
PCG76813.1
AGBW02009250
OWR51255.1
RSAL01000001
RVE55275.1
+ More
KQ459596 KPI95266.1 KQ460118 KPJ17707.1 FX985782 BBA93669.1 NEVH01006756 PNF36443.1 KK853153 KDR10581.1 GFDF01008378 JAV05706.1 GL767674 EFZ13281.1 BABH01028209 BABH01028210 BABH01028211 BABH01028212 ADTU01017499 KQ976453 KYM85435.1 KQ981920 KYN33006.1 KQ982612 KYQ53861.1 GL887813 EGI69950.1 KQ978317 KYM95279.1 KK107293 QOIP01000005 EZA53445.1 RLU22890.1 KQ978579 KYN30011.1 GL448708 EFN83990.1 GBYB01010825 JAG80592.1 KQ414667 KOC64712.1 GL436519 EFN71863.1 NNAY01000904 OXU25996.1 GEZM01096134 JAV55134.1 AAZX01002644 AJWK01029466 KQ435863 KOX70388.1 KZ288194 PBC33915.1 KQ434905 KZC11204.1 LBMM01000453 KMQ98301.1 GAIX01003393 JAA89167.1 KQ971354 EFA07036.1 CH477241 EAT46412.1 DS235219 EEB13380.1 EAT46411.1 GANO01001076 JAB58795.1 UFQT01000097 SSX19784.1 JXUM01102798 JXUM01102799 JXUM01102800 JXUM01102801 KQ564753 KXJ71791.1 GGFK01003593 MBW36914.1 ADMH02001133 ETN63993.1 CVRI01000040 CRK94878.1 GFDL01012001 JAV23044.1 DS232338 EDS40298.1 ATLV01026020 KE525405 KFB52770.1 ODYU01009236 SOQ53509.1 AAAB01008859 EAA08181.2 APCN01000376 AXCM01000492 AXCN02000116 GGFJ01001025 MBW50166.1 APGK01036456 APGK01036457 APGK01036458 KB740940 ENN77681.1 GL732524 EFX89472.1 GDIQ01182249 JAK69476.1 GDIQ01044145 JAN50592.1 GDIQ01145109 GDIQ01061387 GDIQ01034995 JAN33350.1 GDIQ01214033 JAK37692.1 GDIQ01106321 JAL45405.1 GDIQ01068640 JAN26097.1 GDIP01202767 GDIP01030642 JAJ20635.1 KB201304 ESO97964.1 GDIP01234473 GDIQ01110570 JAI88928.1 JAL41156.1 GDIQ01174390 JAK77335.1 GDIQ01095023 JAL56703.1 GDIQ01058842 JAN35895.1 GG666646 EEN46206.1 FR904832 CDQ72425.1 NBAG03000297 PNI45173.1 NDHI03003558 PNJ22128.1 JSUE03012183 JSUE03012184 AK294493 BAH11787.1 AP003037 AP003108 BT058895 ACN10608.1 JW862087 AFO94604.1 AAKN02045307 KB030625 ELK13480.1 AQIA01020060 AQIA01020061 AQIA01020062 AQIA01020063 CABD030079450 CABD030079451 CABD030079452
KQ459596 KPI95266.1 KQ460118 KPJ17707.1 FX985782 BBA93669.1 NEVH01006756 PNF36443.1 KK853153 KDR10581.1 GFDF01008378 JAV05706.1 GL767674 EFZ13281.1 BABH01028209 BABH01028210 BABH01028211 BABH01028212 ADTU01017499 KQ976453 KYM85435.1 KQ981920 KYN33006.1 KQ982612 KYQ53861.1 GL887813 EGI69950.1 KQ978317 KYM95279.1 KK107293 QOIP01000005 EZA53445.1 RLU22890.1 KQ978579 KYN30011.1 GL448708 EFN83990.1 GBYB01010825 JAG80592.1 KQ414667 KOC64712.1 GL436519 EFN71863.1 NNAY01000904 OXU25996.1 GEZM01096134 JAV55134.1 AAZX01002644 AJWK01029466 KQ435863 KOX70388.1 KZ288194 PBC33915.1 KQ434905 KZC11204.1 LBMM01000453 KMQ98301.1 GAIX01003393 JAA89167.1 KQ971354 EFA07036.1 CH477241 EAT46412.1 DS235219 EEB13380.1 EAT46411.1 GANO01001076 JAB58795.1 UFQT01000097 SSX19784.1 JXUM01102798 JXUM01102799 JXUM01102800 JXUM01102801 KQ564753 KXJ71791.1 GGFK01003593 MBW36914.1 ADMH02001133 ETN63993.1 CVRI01000040 CRK94878.1 GFDL01012001 JAV23044.1 DS232338 EDS40298.1 ATLV01026020 KE525405 KFB52770.1 ODYU01009236 SOQ53509.1 AAAB01008859 EAA08181.2 APCN01000376 AXCM01000492 AXCN02000116 GGFJ01001025 MBW50166.1 APGK01036456 APGK01036457 APGK01036458 KB740940 ENN77681.1 GL732524 EFX89472.1 GDIQ01182249 JAK69476.1 GDIQ01044145 JAN50592.1 GDIQ01145109 GDIQ01061387 GDIQ01034995 JAN33350.1 GDIQ01214033 JAK37692.1 GDIQ01106321 JAL45405.1 GDIQ01068640 JAN26097.1 GDIP01202767 GDIP01030642 JAJ20635.1 KB201304 ESO97964.1 GDIP01234473 GDIQ01110570 JAI88928.1 JAL41156.1 GDIQ01174390 JAK77335.1 GDIQ01095023 JAL56703.1 GDIQ01058842 JAN35895.1 GG666646 EEN46206.1 FR904832 CDQ72425.1 NBAG03000297 PNI45173.1 NDHI03003558 PNJ22128.1 JSUE03012183 JSUE03012184 AK294493 BAH11787.1 AP003037 AP003108 BT058895 ACN10608.1 JW862087 AFO94604.1 AAKN02045307 KB030625 ELK13480.1 AQIA01020060 AQIA01020061 AQIA01020062 AQIA01020063 CABD030079450 CABD030079451 CABD030079452
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000235965
+ More
UP000027135 UP000005204 UP000005205 UP000078540 UP000078541 UP000075809 UP000007755 UP000078542 UP000053097 UP000279307 UP000078492 UP000008237 UP000053825 UP000000311 UP000215335 UP000002358 UP000092461 UP000053105 UP000242457 UP000005203 UP000076502 UP000192223 UP000036403 UP000007266 UP000008820 UP000009046 UP000069940 UP000249989 UP000000673 UP000183832 UP000069272 UP000002320 UP000030765 UP000076408 UP000075884 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075920 UP000075881 UP000075883 UP000075886 UP000075885 UP000085678 UP000019118 UP000075880 UP000000305 UP000075900 UP000030746 UP000001554 UP000193380 UP000233180 UP000006718 UP000005640 UP000087266 UP000265140 UP000002281 UP000005447 UP000233120 UP000010552 UP000233100 UP000001519
UP000027135 UP000005204 UP000005205 UP000078540 UP000078541 UP000075809 UP000007755 UP000078542 UP000053097 UP000279307 UP000078492 UP000008237 UP000053825 UP000000311 UP000215335 UP000002358 UP000092461 UP000053105 UP000242457 UP000005203 UP000076502 UP000192223 UP000036403 UP000007266 UP000008820 UP000009046 UP000069940 UP000249989 UP000000673 UP000183832 UP000069272 UP000002320 UP000030765 UP000076408 UP000075884 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075920 UP000075881 UP000075883 UP000075886 UP000075885 UP000085678 UP000019118 UP000075880 UP000000305 UP000075900 UP000030746 UP000001554 UP000193380 UP000233180 UP000006718 UP000005640 UP000087266 UP000265140 UP000002281 UP000005447 UP000233120 UP000010552 UP000233100 UP000001519
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JZ82
A0A212FBZ7
A0A3S2LJK0
A0A194PP99
A0A194RJH7
A0A2Z5TRE2
+ More
A0A2J7R6I4 A0A067QWW9 A0A1L8DGY1 E9J1W0 H9J5F0 A0A158NJ58 A0A195BL46 A0A195EXY0 A0A151X0H4 F4W766 A0A195C340 A0A026WBL0 A0A151JS91 E2BJY1 A0A0C9RQP9 A0A0L7R1I9 E2A411 A0A232F584 A0A1Y1K0V9 K7IUN5 A0A1B0CUN8 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A154PI86 A0A1W4X581 A0A0J7L6L1 S4PYF2 D6WR91 Q17I86 E0VJ24 Q17I87 U5EVD6 A0A1S4F1P1 A0A336LNQ7 A0A182H165 A0A2M4A7Y9 W5JL18 A0A1J1I7F1 A0A182FT49 A0A1Q3F644 B0X4E5 A0A084WRC6 A0A182XWW1 A0A182NLA8 A0A2H1WKD0 A0A182UN77 A0A182UGX6 A0A182LF28 A0A182XI06 Q7QC66 A0A182HSY7 A0A182VXE5 A0A182JRI8 A0A182MI35 A0A182QID4 A0A182PAF4 A0A1S3IIM7 A0A2M4BAV2 N6UAL4 A0A182JC38 A0A1S3IIX8 E9FU30 A0A182R366 A0A0P5KLL0 A0A0P6GFZ4 A0A0P6FRV4 A0A0P5IFL8 A0A0P5RR17 A0A0P6E5N7 A0A0P5AAI7 V4C8B8 A0A0N7ZL83 A0A0P5LWX0 A0A0P5RXC9 A0A0P6FCL2 C3ZMJ6 A0A060X6D7 A0A2K6L728 A0A2J8LD23 A0A2J8SMZ1 F7HN48 B7Z2A1 F5GY55 C0H989 A0A3P8Z8J6 F6QDR0 V9K9R8 A0A286XPJ3 A0A2K6BUZ0 A0A2K6L717 L5KQI8 A0A2K5VZB4 G3QN74
A0A2J7R6I4 A0A067QWW9 A0A1L8DGY1 E9J1W0 H9J5F0 A0A158NJ58 A0A195BL46 A0A195EXY0 A0A151X0H4 F4W766 A0A195C340 A0A026WBL0 A0A151JS91 E2BJY1 A0A0C9RQP9 A0A0L7R1I9 E2A411 A0A232F584 A0A1Y1K0V9 K7IUN5 A0A1B0CUN8 A0A0N0BDG5 A0A2A3EQ74 A0A087ZZR8 A0A154PI86 A0A1W4X581 A0A0J7L6L1 S4PYF2 D6WR91 Q17I86 E0VJ24 Q17I87 U5EVD6 A0A1S4F1P1 A0A336LNQ7 A0A182H165 A0A2M4A7Y9 W5JL18 A0A1J1I7F1 A0A182FT49 A0A1Q3F644 B0X4E5 A0A084WRC6 A0A182XWW1 A0A182NLA8 A0A2H1WKD0 A0A182UN77 A0A182UGX6 A0A182LF28 A0A182XI06 Q7QC66 A0A182HSY7 A0A182VXE5 A0A182JRI8 A0A182MI35 A0A182QID4 A0A182PAF4 A0A1S3IIM7 A0A2M4BAV2 N6UAL4 A0A182JC38 A0A1S3IIX8 E9FU30 A0A182R366 A0A0P5KLL0 A0A0P6GFZ4 A0A0P6FRV4 A0A0P5IFL8 A0A0P5RR17 A0A0P6E5N7 A0A0P5AAI7 V4C8B8 A0A0N7ZL83 A0A0P5LWX0 A0A0P5RXC9 A0A0P6FCL2 C3ZMJ6 A0A060X6D7 A0A2K6L728 A0A2J8LD23 A0A2J8SMZ1 F7HN48 B7Z2A1 F5GY55 C0H989 A0A3P8Z8J6 F6QDR0 V9K9R8 A0A286XPJ3 A0A2K6BUZ0 A0A2K6L717 L5KQI8 A0A2K5VZB4 G3QN74
PDB
4A0B
E-value=0,
Score=3316
Ontologies
GO
GO:0006281
GO:0003676
GO:0005634
GO:0003684
GO:0043161
GO:0022857
GO:0016021
GO:0005525
GO:0071987
GO:1902188
GO:0043066
GO:0070914
GO:0005654
GO:0005737
GO:0031464
GO:0035518
GO:0016055
GO:0042752
GO:0031465
GO:0045722
GO:0045070
GO:1901990
GO:0097602
GO:0046726
GO:0030674
GO:0045732
GO:0044877
GO:0051702
GO:0003700
GO:0008270
GO:0043565
GO:0003677
GO:0003707
GO:0006259
GO:0005515
PANTHER
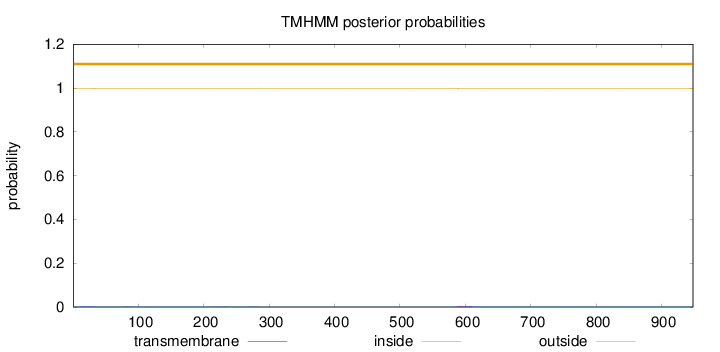
Topology
Length:
948
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.128230000000001
Exp number, first 60 AAs:
0.03685
Total prob of N-in:
0.00279
outside
1 - 948
Population Genetic Test Statistics
Pi
216.508393
Theta
173.694665
Tajima's D
0.259856
CLR
1135.433459
CSRT
0.44312784360782
Interpretation
Uncertain
