Pre Gene Modal
BGIBMGA004739
Annotation
PREDICTED:_b(0?+)-type_amino_acid_transporter_1_isoform_X1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.989
Sequence
CDS
ATGTCCGCTCAAAAAGCGGAAATATCTGATAGACCTGAAACTCTAAGCAGACAGCACAGCGAATGCGGCAGCGCAGTAAAATTAAAACGAGAACTCGGCTTGTTCTCCGCTGTTAACCTAATTCTTGGGGTAATGATTGGATCTGGTATATTCGTTTCCCCAGCTTCAGCATTGGAGTATTCTGGGTCCGTAGCGCTATGTCTAATAATATGGACAGTTTCAGGAATAATATCCTTACTAGGAGCATTATCGTTCGCTGAGTTAGGAACTGTAGTTGGAAAATCAGGAGCCGAGTATGCATATTTTCAGGAAGCCTTTGGGAAAATTCACAAATTTTGGGGTCCATTACCGTCTTTCTCATGTGCTTGGATCTATGTAATGATCCTTAGACCGGCAGAAGTAGCTATTGTGGTAATGACATTCGCCGAATATGCAATTCAGCCAATTACCATTGATGTACATCCAGATTATAAAGACAATGCCATCAAACTAGCATCACTCGCAGCCTTGTTCATCATGACCTACATCAACATAACTAGCGTCAAATTGTTCGTGAAGGTACAGAACGTCTTTGGAGTGTGTAAAGTTCTAGCGTGCCTCGTCGTCATTGGTGGAGGCATCTACGAAATCGCGAAAGGCAACACCTCTCATTTAAATAAAGGTTTTGAGGGTAGCACAACTAATCCCGGAAGTATTGCTCTGTCTCTGTACTACGGGCTATGGGCATACGACGGCTGGAACAGCGTGACCGTTGTCACTGAGGAAATCATTAATCCTGGCGTAAATGTTCCGTTGAGCATATCAATCGCCGTGCCGCTCATCACGGCGCTATACGTTTTCATGAACGTTGCCTATATGACGGTGCTGAGCTACGCGGAAATGACGTCCGGCACTGCGGTCGCGGTGGCGTTCGGCGCAAGAGTACTCGGGCCGTTTAGCTTCATCATGCCGCTCGGTGTTGCCGTGGCAACATTTGGCTGCGCTATGAGTGTGCAGTTTGGTGTTACCAGGGTGTGTTACACAGCGGCTCGTGGGGGTCACATGCTTGAGCTTTTCTCGTATGTCAACATAAAACGTCTTACACCCGCGCCGGCTGTGGCATTTCAAGCCTTCTTAACAGCTATATTCATATCGGTCGGAAACATCAAAACTTTAATACAGTTCGCTAGTTGGTTCCTGTGGTTTTTCTACGGTCTCGCGATGGTGGCGTTGTTGGTGCTCAGGAAGACTCAAGCCGCTAAACCACGTCCGTATAGAGTGCCAACGCCGATACCGGTTTTCGTGCTGATTGTGGCCATATTCCTATCGGTCCTGCCCATAGTTGAAGACCCTTCCGTGAAATATCTAATGGCCATAGGATTCATTCTGTTAGGCGTCGTCGTGTACACTGTATTTGTGTATTACAAGAGAACTCCGTCGGCTTTAATAAGAAAACTCACGTTTGTCACTCAAATTTTATTCGAAAGCGTACCACCGAGCAAGGATCAAGATGACTGA
Protein
MSAQKAEISDRPETLSRQHSECGSAVKLKRELGLFSAVNLILGVMIGSGIFVSPASALEYSGSVALCLIIWTVSGIISLLGALSFAELGTVVGKSGAEYAYFQEAFGKIHKFWGPLPSFSCAWIYVMILRPAEVAIVVMTFAEYAIQPITIDVHPDYKDNAIKLASLAALFIMTYINITSVKLFVKVQNVFGVCKVLACLVVIGGGIYEIAKGNTSHLNKGFEGSTTNPGSIALSLYYGLWAYDGWNSVTVVTEEIINPGVNVPLSISIAVPLITALYVFMNVAYMTVLSYAEMTSGTAVAVAFGARVLGPFSFIMPLGVAVATFGCAMSVQFGVTRVCYTAARGGHMLELFSYVNIKRLTPAPAVAFQAFLTAIFISVGNIKTLIQFASWFLWFFYGLAMVALLVLRKTQAAKPRPYRVPTPIPVFVLIVAIFLSVLPIVEDPSVKYLMAIGFILLGVVVYTVFVYYKRTPSALIRKLTFVTQILFESVPPSKDQDD
Summary
Uniprot
A0A2A4JZI4
A0A1B0GGZ1
A0A1B0DPF4
A0A1L8DEK8
U5EKX7
D6WEE2
+ More
A0A182HWR7 Q7Q6V2 A0A182NGX1 A0A182UXX3 A0A182LJ30 A0A084W475 A0A182XMW1 A0A1J1IUV0 A0A182H3B3 A0A182PHN1 A0A182RFA2 Q16YX2 A0A182WIH6 A0A182Y1A9 A0A182K3R6 A0A182MBR7 A0A1Q3FNL6 A0A2M4AS29 A0A2M4ARW5 A0A182Q897 W5JFA2 A0A182J4Q0 A0A067R571 B0X5U9 A0A182FHY5 A0A2J7RSD2 A0A1Q3FNN4 A0A1Y1L9E7 A0A182U190 A0A2P8Y8D8 A0A336M139 A0A067REX2 J9JMD4 A0A1L8DE74 N6TV61 A0A2J7QWV5 E2BVQ4 U4U9S4 A0A1B6EGJ5 A0A1B6E6U7 A0A0J7NLP3 A0A336LV27 A0A069DTW8 A0A1B6FK86 A0A0P4VNR9 A0A023F3U0 E2AWL9 A0A026W5Z2 A0A2J7QMN0 A0A1B6KY22 F4X544 A0A151XFX2 A0A1B6FDB9 A0A195FNR9 A0A158NQ08 A0A195BI05 A0A1B6L852 A0A232FP37 A0A154NZD2 A0A0L7QZN3 A0A0M9A5L1 A0A0A9ZD36 A0A0K8T8G6 A0A195CUG0 A0A2J7QMP6 A0A1B6C7D7 A0A151IVX8 A0A2J7QMR0 A0A2S2R0U7 A0A067RHI4 A0A1B6IL33 A0A2A3E3M1 A0A088AU01 A0A2J7QMP0 A0A310SNF5 E9IEU4 A0A1B6HRY7 A0A2S2QJ57 A0A2S2R3C8 A0A2S2Q744 A0A2M4CQD6 T1E9J8 T1E9K0 A0A2J7QMS5 A0A067QUR4 A0A0A9Z6P3 A0A146M5U7 A0A0A9Z174 A0A146L4D7 A0A0P5IU45
A0A182HWR7 Q7Q6V2 A0A182NGX1 A0A182UXX3 A0A182LJ30 A0A084W475 A0A182XMW1 A0A1J1IUV0 A0A182H3B3 A0A182PHN1 A0A182RFA2 Q16YX2 A0A182WIH6 A0A182Y1A9 A0A182K3R6 A0A182MBR7 A0A1Q3FNL6 A0A2M4AS29 A0A2M4ARW5 A0A182Q897 W5JFA2 A0A182J4Q0 A0A067R571 B0X5U9 A0A182FHY5 A0A2J7RSD2 A0A1Q3FNN4 A0A1Y1L9E7 A0A182U190 A0A2P8Y8D8 A0A336M139 A0A067REX2 J9JMD4 A0A1L8DE74 N6TV61 A0A2J7QWV5 E2BVQ4 U4U9S4 A0A1B6EGJ5 A0A1B6E6U7 A0A0J7NLP3 A0A336LV27 A0A069DTW8 A0A1B6FK86 A0A0P4VNR9 A0A023F3U0 E2AWL9 A0A026W5Z2 A0A2J7QMN0 A0A1B6KY22 F4X544 A0A151XFX2 A0A1B6FDB9 A0A195FNR9 A0A158NQ08 A0A195BI05 A0A1B6L852 A0A232FP37 A0A154NZD2 A0A0L7QZN3 A0A0M9A5L1 A0A0A9ZD36 A0A0K8T8G6 A0A195CUG0 A0A2J7QMP6 A0A1B6C7D7 A0A151IVX8 A0A2J7QMR0 A0A2S2R0U7 A0A067RHI4 A0A1B6IL33 A0A2A3E3M1 A0A088AU01 A0A2J7QMP0 A0A310SNF5 E9IEU4 A0A1B6HRY7 A0A2S2QJ57 A0A2S2R3C8 A0A2S2Q744 A0A2M4CQD6 T1E9J8 T1E9K0 A0A2J7QMS5 A0A067QUR4 A0A0A9Z6P3 A0A146M5U7 A0A0A9Z174 A0A146L4D7 A0A0P5IU45
Pubmed
EMBL
NWSH01000383
PCG76802.1
AJWK01000973
AJWK01000974
AJWK01000975
AJVK01008079
+ More
AJVK01008080 AJVK01008081 GFDF01009216 JAV04868.1 GANO01001652 JAB58219.1 KQ971318 EFA00361.2 APCN01001533 AAAB01008960 EAA11727.4 ATLV01020284 KE525297 KFB45019.1 CVRI01000058 CRL02918.1 JXUM01024456 JXUM01024457 JXUM01024458 JXUM01024459 CH477507 CH477233 EAT39815.1 EAT46790.1 AXCM01002623 GFDL01005957 JAV29088.1 GGFK01010266 MBW43587.1 GGFK01010225 MBW43546.1 AXCN02000361 ADMH02001577 ETN62013.1 KK852694 KDR18305.1 DS232396 EDS41080.1 NEVH01000261 PNF43732.1 GFDL01005952 JAV29093.1 GEZM01066079 JAV68206.1 PYGN01000807 PSN40526.1 UFQS01000305 UFQT01000305 SSX02667.1 SSX23041.1 KK852680 KDR18609.1 ABLF02036152 GFDF01009407 JAV04677.1 APGK01050872 KB741178 ENN73155.1 NEVH01009418 PNF33059.1 GL450924 EFN80237.1 KB632256 ERL90679.1 GEDC01000263 JAS37035.1 GEDC01003645 JAS33653.1 LBMM01003549 KMQ93395.1 UFQT01000212 SSX21794.1 GBGD01001346 JAC87543.1 GECZ01019143 JAS50626.1 GDKW01001993 JAI54602.1 GBBI01003123 JAC15589.1 GL443346 EFN62183.1 KK107388 QOIP01000001 EZA51492.1 RLU26761.1 NEVH01013204 PNF29854.1 GEBQ01023823 JAT16154.1 GL888687 EGI58438.1 KQ982182 KYQ59241.1 GECZ01021582 JAS48187.1 KQ981382 KYN42061.1 ADTU01022843 ADTU01022844 KQ976464 KYM84483.1 GEBQ01020088 JAT19889.1 NNAY01000002 OXU32289.1 KQ434783 KZC04942.1 KQ414676 KOC64084.1 KQ435729 KOX77700.1 GBHO01000462 GBHO01000461 JAG43142.1 JAG43143.1 GBRD01004140 JAG61681.1 KQ977279 KYN04318.1 PNF29860.1 GEDC01027900 JAS09398.1 KQ980880 KYN12011.1 PNF29858.1 GGMS01014448 MBY83651.1 KDR18608.1 GECU01020080 JAS87626.1 KZ288721 PBC26084.1 PNF29861.1 KQ762864 OAD55398.1 GL762722 EFZ20910.1 GECU01030280 JAS77426.1 GGMS01008347 MBY77550.1 GGMS01015313 MBY84516.1 GGMS01004208 MBY73411.1 GGFL01002920 MBW67098.1 GAMD01001788 JAA99802.1 GAMD01001783 JAA99807.1 PNF29853.1 KK852927 KDR13693.1 GBHO01004576 JAG39028.1 GDHC01003491 JAQ15138.1 GBHO01004577 JAG39027.1 GDHC01016989 JAQ01640.1 GDIP01233336 GDIQ01209800 JAI90065.1 JAK41925.1
AJVK01008080 AJVK01008081 GFDF01009216 JAV04868.1 GANO01001652 JAB58219.1 KQ971318 EFA00361.2 APCN01001533 AAAB01008960 EAA11727.4 ATLV01020284 KE525297 KFB45019.1 CVRI01000058 CRL02918.1 JXUM01024456 JXUM01024457 JXUM01024458 JXUM01024459 CH477507 CH477233 EAT39815.1 EAT46790.1 AXCM01002623 GFDL01005957 JAV29088.1 GGFK01010266 MBW43587.1 GGFK01010225 MBW43546.1 AXCN02000361 ADMH02001577 ETN62013.1 KK852694 KDR18305.1 DS232396 EDS41080.1 NEVH01000261 PNF43732.1 GFDL01005952 JAV29093.1 GEZM01066079 JAV68206.1 PYGN01000807 PSN40526.1 UFQS01000305 UFQT01000305 SSX02667.1 SSX23041.1 KK852680 KDR18609.1 ABLF02036152 GFDF01009407 JAV04677.1 APGK01050872 KB741178 ENN73155.1 NEVH01009418 PNF33059.1 GL450924 EFN80237.1 KB632256 ERL90679.1 GEDC01000263 JAS37035.1 GEDC01003645 JAS33653.1 LBMM01003549 KMQ93395.1 UFQT01000212 SSX21794.1 GBGD01001346 JAC87543.1 GECZ01019143 JAS50626.1 GDKW01001993 JAI54602.1 GBBI01003123 JAC15589.1 GL443346 EFN62183.1 KK107388 QOIP01000001 EZA51492.1 RLU26761.1 NEVH01013204 PNF29854.1 GEBQ01023823 JAT16154.1 GL888687 EGI58438.1 KQ982182 KYQ59241.1 GECZ01021582 JAS48187.1 KQ981382 KYN42061.1 ADTU01022843 ADTU01022844 KQ976464 KYM84483.1 GEBQ01020088 JAT19889.1 NNAY01000002 OXU32289.1 KQ434783 KZC04942.1 KQ414676 KOC64084.1 KQ435729 KOX77700.1 GBHO01000462 GBHO01000461 JAG43142.1 JAG43143.1 GBRD01004140 JAG61681.1 KQ977279 KYN04318.1 PNF29860.1 GEDC01027900 JAS09398.1 KQ980880 KYN12011.1 PNF29858.1 GGMS01014448 MBY83651.1 KDR18608.1 GECU01020080 JAS87626.1 KZ288721 PBC26084.1 PNF29861.1 KQ762864 OAD55398.1 GL762722 EFZ20910.1 GECU01030280 JAS77426.1 GGMS01008347 MBY77550.1 GGMS01015313 MBY84516.1 GGMS01004208 MBY73411.1 GGFL01002920 MBW67098.1 GAMD01001788 JAA99802.1 GAMD01001783 JAA99807.1 PNF29853.1 KK852927 KDR13693.1 GBHO01004576 JAG39028.1 GDHC01003491 JAQ15138.1 GBHO01004577 JAG39027.1 GDHC01016989 JAQ01640.1 GDIP01233336 GDIQ01209800 JAI90065.1 JAK41925.1
Proteomes
UP000218220
UP000092461
UP000092462
UP000007266
UP000075840
UP000007062
+ More
UP000075884 UP000075903 UP000075882 UP000030765 UP000076407 UP000183832 UP000069940 UP000075885 UP000075900 UP000008820 UP000075920 UP000076408 UP000075881 UP000075883 UP000075886 UP000000673 UP000075880 UP000027135 UP000002320 UP000069272 UP000235965 UP000075902 UP000245037 UP000007819 UP000019118 UP000008237 UP000030742 UP000036403 UP000000311 UP000053097 UP000279307 UP000007755 UP000075809 UP000078541 UP000005205 UP000078540 UP000215335 UP000076502 UP000053825 UP000053105 UP000078542 UP000078492 UP000242457 UP000005203
UP000075884 UP000075903 UP000075882 UP000030765 UP000076407 UP000183832 UP000069940 UP000075885 UP000075900 UP000008820 UP000075920 UP000076408 UP000075881 UP000075883 UP000075886 UP000000673 UP000075880 UP000027135 UP000002320 UP000069272 UP000235965 UP000075902 UP000245037 UP000007819 UP000019118 UP000008237 UP000030742 UP000036403 UP000000311 UP000053097 UP000279307 UP000007755 UP000075809 UP000078541 UP000005205 UP000078540 UP000215335 UP000076502 UP000053825 UP000053105 UP000078542 UP000078492 UP000242457 UP000005203
Pfam
PF13520 AA_permease_2
Interpro
IPR002293
AA/rel_permease1
ProteinModelPortal
A0A2A4JZI4
A0A1B0GGZ1
A0A1B0DPF4
A0A1L8DEK8
U5EKX7
D6WEE2
+ More
A0A182HWR7 Q7Q6V2 A0A182NGX1 A0A182UXX3 A0A182LJ30 A0A084W475 A0A182XMW1 A0A1J1IUV0 A0A182H3B3 A0A182PHN1 A0A182RFA2 Q16YX2 A0A182WIH6 A0A182Y1A9 A0A182K3R6 A0A182MBR7 A0A1Q3FNL6 A0A2M4AS29 A0A2M4ARW5 A0A182Q897 W5JFA2 A0A182J4Q0 A0A067R571 B0X5U9 A0A182FHY5 A0A2J7RSD2 A0A1Q3FNN4 A0A1Y1L9E7 A0A182U190 A0A2P8Y8D8 A0A336M139 A0A067REX2 J9JMD4 A0A1L8DE74 N6TV61 A0A2J7QWV5 E2BVQ4 U4U9S4 A0A1B6EGJ5 A0A1B6E6U7 A0A0J7NLP3 A0A336LV27 A0A069DTW8 A0A1B6FK86 A0A0P4VNR9 A0A023F3U0 E2AWL9 A0A026W5Z2 A0A2J7QMN0 A0A1B6KY22 F4X544 A0A151XFX2 A0A1B6FDB9 A0A195FNR9 A0A158NQ08 A0A195BI05 A0A1B6L852 A0A232FP37 A0A154NZD2 A0A0L7QZN3 A0A0M9A5L1 A0A0A9ZD36 A0A0K8T8G6 A0A195CUG0 A0A2J7QMP6 A0A1B6C7D7 A0A151IVX8 A0A2J7QMR0 A0A2S2R0U7 A0A067RHI4 A0A1B6IL33 A0A2A3E3M1 A0A088AU01 A0A2J7QMP0 A0A310SNF5 E9IEU4 A0A1B6HRY7 A0A2S2QJ57 A0A2S2R3C8 A0A2S2Q744 A0A2M4CQD6 T1E9J8 T1E9K0 A0A2J7QMS5 A0A067QUR4 A0A0A9Z6P3 A0A146M5U7 A0A0A9Z174 A0A146L4D7 A0A0P5IU45
A0A182HWR7 Q7Q6V2 A0A182NGX1 A0A182UXX3 A0A182LJ30 A0A084W475 A0A182XMW1 A0A1J1IUV0 A0A182H3B3 A0A182PHN1 A0A182RFA2 Q16YX2 A0A182WIH6 A0A182Y1A9 A0A182K3R6 A0A182MBR7 A0A1Q3FNL6 A0A2M4AS29 A0A2M4ARW5 A0A182Q897 W5JFA2 A0A182J4Q0 A0A067R571 B0X5U9 A0A182FHY5 A0A2J7RSD2 A0A1Q3FNN4 A0A1Y1L9E7 A0A182U190 A0A2P8Y8D8 A0A336M139 A0A067REX2 J9JMD4 A0A1L8DE74 N6TV61 A0A2J7QWV5 E2BVQ4 U4U9S4 A0A1B6EGJ5 A0A1B6E6U7 A0A0J7NLP3 A0A336LV27 A0A069DTW8 A0A1B6FK86 A0A0P4VNR9 A0A023F3U0 E2AWL9 A0A026W5Z2 A0A2J7QMN0 A0A1B6KY22 F4X544 A0A151XFX2 A0A1B6FDB9 A0A195FNR9 A0A158NQ08 A0A195BI05 A0A1B6L852 A0A232FP37 A0A154NZD2 A0A0L7QZN3 A0A0M9A5L1 A0A0A9ZD36 A0A0K8T8G6 A0A195CUG0 A0A2J7QMP6 A0A1B6C7D7 A0A151IVX8 A0A2J7QMR0 A0A2S2R0U7 A0A067RHI4 A0A1B6IL33 A0A2A3E3M1 A0A088AU01 A0A2J7QMP0 A0A310SNF5 E9IEU4 A0A1B6HRY7 A0A2S2QJ57 A0A2S2R3C8 A0A2S2Q744 A0A2M4CQD6 T1E9J8 T1E9K0 A0A2J7QMS5 A0A067QUR4 A0A0A9Z6P3 A0A146M5U7 A0A0A9Z174 A0A146L4D7 A0A0P5IU45
PDB
6IRT
E-value=5.45315e-74,
Score=707
Ontologies
GO
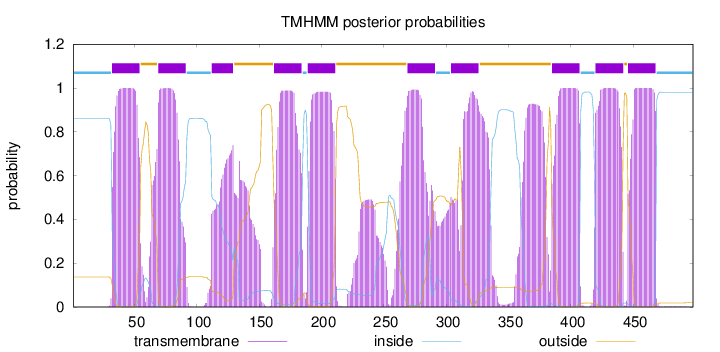
Topology
Length:
498
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
255.26598
Exp number, first 60 AAs:
22.07906
Total prob of N-in:
0.86259
POSSIBLE N-term signal
sequence
inside
1 - 31
TMhelix
32 - 54
outside
55 - 68
TMhelix
69 - 91
inside
92 - 111
TMhelix
112 - 129
outside
130 - 161
TMhelix
162 - 184
inside
185 - 188
TMhelix
189 - 211
outside
212 - 268
TMhelix
269 - 291
inside
292 - 303
TMhelix
304 - 326
outside
327 - 384
TMhelix
385 - 407
inside
408 - 419
TMhelix
420 - 442
outside
443 - 445
TMhelix
446 - 468
inside
469 - 498
Population Genetic Test Statistics
Pi
232.718637
Theta
194.520491
Tajima's D
0.603302
CLR
0.393572
CSRT
0.541872906354682
Interpretation
Uncertain
