Gene
KWMTBOMO15419 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004776
Annotation
PREDICTED:_protein_arginine_N-methyltransferase_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.736
Sequence
CDS
ATGGAAACAATGGACGTAGCTCAAGAAGGAACATCGACGGTTTCAACGCCTGTTGTGGAAAATGGACCTGACAAGAATGTTACTGCCGAAGAAATGACGTCGAGGGATTATTATTTCGACTCATATGCACATTTCGGCATCCACGAGGAAATGTTAAAAGATGAAGTCCGCACTCTCACCTACAAAAATGCCATGTACCACAATAAGCATTTATTTCAAGGAAAGACAGTCCTCGATATAGGCTGTGGAACTGGGATCCTGTCGATGTTTGCAGCTAAAGCAGGTGCTACCAAGGTCTTAGCGATAGAGTGCTCCAACATTGTTGACTATGCTCGTAAAATTATTGAGGCAAACAGACTCGATGATGTCATTGAGATTATTAAAGGAAAGGTGGAGGAAGTTGAATTACCAGTTGACAAAGTGGACATCATAATATCAGAATGGATGGGCTACTGTCTATTCTATGAGAGTATGCTTGATACGGTGCTATATGCCAGAGATAAGTGGCTCAAGCCAGATGGGATGATGTTTCCAGATAGATGTACATTATTTATATGTGGTATAGAAGATCGGCAATACAAAGATGAGAAGATCAACTGGTGGGATGATGTGTACGGCTTTGACATGTCTTCAATCAGGAAGGTGGCTATATCTGAACCCCTTGTAGATGTGGTTGATGCTAAACAGGTTGTCACGAATTCATCCTTATTAAAAGAAATAGATTTATACACAGTCAAGAAAGAGGATCTTAATTTTGAATCAAAATTCCATTTGCATGTCCGTCGGAACGACTTTATCCAAGCCTTAGTGACGTACTTCAACGTTGAGTTCACGAAGTCACACAAACGTCTCGGGTTCAGTACCGCGCCGGACGCTCCCTACACGCACTGGAAGCAGACAGTCTTCTACTTTGATGATTTTATGACGGTAAAGAAATTCGAAGAGATAACCGGTACATTCTCGATGCGCCAGAACGCAAGGAACAATCGCGACTTGGACTTTGAGATCGAGATCGACTTCAAGGGGGAACTCTGTCAGGTCAACGAGAAGAATCACTACAGGATGCGCTAG
Protein
METMDVAQEGTSTVSTPVVENGPDKNVTAEEMTSRDYYFDSYAHFGIHEEMLKDEVRTLTYKNAMYHNKHLFQGKTVLDIGCGTGILSMFAAKAGATKVLAIECSNIVDYARKIIEANRLDDVIEIIKGKVEEVELPVDKVDIIISEWMGYCLFYESMLDTVLYARDKWLKPDGMMFPDRCTLFICGIEDRQYKDEKINWWDDVYGFDMSSIRKVAISEPLVDVVDAKQVVTNSSLLKEIDLYTVKKEDLNFESKFHLHVRRNDFIQALVTYFNVEFTKSHKRLGFSTAPDAPYTHWKQTVFYFDDFMTVKKFEEITGTFSMRQNARNNRDLDFEIEIDFKGELCQVNEKNHYRMR
Summary
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. Protein arginine N-methyltransferase family.
Uniprot
H9J5I6
A0A2A4JZ74
A0A3S2P291
A0A2H1V2N3
E3UKI5
A0A0K0PYY8
+ More
A0A194PR89 S4P7D1 A0A212EXI1 A0A2P8YHX0 A0A182V6T4 A0A182KX41 A0A182TPM7 A0A182XK30 Q7PRE6 A0A182HJW1 A0A182PN09 A0A182NKY4 A0A182QNV1 A0A1B6LU91 A0A0T6B8E5 A0A182R846 A0A182YM54 A0A182T1Q6 A0A195EKC3 F4WC44 A0A195FQP0 A0A1B6GVP5 A0A158NA18 A0A067QQH5 B0W9B5 A0A151WXA2 A0A182M8C9 A0A182VS92 A0A3L8DA14 E9J265 A0A1B6LJG4 A0A2J7QLL8 A0A2M3Z313 E2BZ08 A0A2M4AB66 A0A026WIQ2 A0A2J7QLM3 A0A1B6FJ66 A0A0J7NIZ7 A0A182K728 W5JCN8 A0A154PHI8 A0A084VJP5 E2A2D3 A0A1Q3EWJ4 A0A1Q3EWI3 A0A2M4BRM9 T1DH59 E0VIG1 D6WU94 A0A182IWC5 A0A182GDW4 A0A1B6EFM7 Q17G76 A0A1S4F430 A0A1W4WQZ3 A0A0M9A4F1 A0A2A3EDK7 A0A232F014 A0A1Y1N6P3 J3JVG0 A0A1B0C9H2 N6U8X7 A0A0C9QVA8 U5ETH4 A0A0K8TM86 A0A1L8DLZ2 A0A1L8DLX2 B4PLD3 Q9VGW7 B4QUP6 A0A1W4W0T7 A0ANP8 B4NJY2 A0ANQ8 A0A3B0JLV3 B3P1K1 A0A3B0JLL6 B4M5I2 T1E306 W8CEE4 B4KC99 C0MJ37 A0A087UPY2 A0A034WVH2 A0A0K8WIY1 Q297G9 B4GE03 A0A0L0BQ09 A0A0A1XSZ8 B3M290 B4JHM5 A0A293MW14 A0A1L8EBY0
A0A194PR89 S4P7D1 A0A212EXI1 A0A2P8YHX0 A0A182V6T4 A0A182KX41 A0A182TPM7 A0A182XK30 Q7PRE6 A0A182HJW1 A0A182PN09 A0A182NKY4 A0A182QNV1 A0A1B6LU91 A0A0T6B8E5 A0A182R846 A0A182YM54 A0A182T1Q6 A0A195EKC3 F4WC44 A0A195FQP0 A0A1B6GVP5 A0A158NA18 A0A067QQH5 B0W9B5 A0A151WXA2 A0A182M8C9 A0A182VS92 A0A3L8DA14 E9J265 A0A1B6LJG4 A0A2J7QLL8 A0A2M3Z313 E2BZ08 A0A2M4AB66 A0A026WIQ2 A0A2J7QLM3 A0A1B6FJ66 A0A0J7NIZ7 A0A182K728 W5JCN8 A0A154PHI8 A0A084VJP5 E2A2D3 A0A1Q3EWJ4 A0A1Q3EWI3 A0A2M4BRM9 T1DH59 E0VIG1 D6WU94 A0A182IWC5 A0A182GDW4 A0A1B6EFM7 Q17G76 A0A1S4F430 A0A1W4WQZ3 A0A0M9A4F1 A0A2A3EDK7 A0A232F014 A0A1Y1N6P3 J3JVG0 A0A1B0C9H2 N6U8X7 A0A0C9QVA8 U5ETH4 A0A0K8TM86 A0A1L8DLZ2 A0A1L8DLX2 B4PLD3 Q9VGW7 B4QUP6 A0A1W4W0T7 A0ANP8 B4NJY2 A0ANQ8 A0A3B0JLV3 B3P1K1 A0A3B0JLL6 B4M5I2 T1E306 W8CEE4 B4KC99 C0MJ37 A0A087UPY2 A0A034WVH2 A0A0K8WIY1 Q297G9 B4GE03 A0A0L0BQ09 A0A0A1XSZ8 B3M290 B4JHM5 A0A293MW14 A0A1L8EBY0
Pubmed
19121390
26354079
23622113
22118469
29403074
20966253
+ More
12364791 14747013 17210077 25244985 21719571 21347285 24845553 30249741 21282665 20798317 24508170 20920257 23761445 24438588 20566863 18362917 19820115 26483478 17510324 28648823 28004739 22516182 23537049 26369729 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 16951084 17569856 17569867 19126864 26109357 26109356 24330624 24495485 25348373 15632085 26108605 25830018
12364791 14747013 17210077 25244985 21719571 21347285 24845553 30249741 21282665 20798317 24508170 20920257 23761445 24438588 20566863 18362917 19820115 26483478 17510324 28648823 28004739 22516182 23537049 26369729 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 16951084 17569856 17569867 19126864 26109357 26109356 24330624 24495485 25348373 15632085 26108605 25830018
EMBL
BABH01028218
NWSH01000383
PCG76803.1
RSAL01000001
RVE55273.1
ODYU01000390
+ More
SOQ35091.1 HM449859 ADO32994.1 KP735964 AKQ99124.1 KQ459596 KPI95264.1 GAIX01006431 JAA86129.1 AGBW02011765 OWR46171.1 PYGN01000582 PSN43857.1 AAAB01008851 EAA07364.5 APCN01002209 AXCN02000107 GEBQ01012733 JAT27244.1 LJIG01009176 KRT83604.1 KQ978739 KYN28715.1 GL888070 EGI68024.1 KQ981305 KYN42900.1 GECZ01003245 JAS66524.1 ADTU01009939 KK853567 KDR06666.1 DS231863 EDS39975.1 KQ982686 KYQ52295.1 AXCM01004252 QOIP01000011 RLU17174.1 GL767819 EFZ13091.1 GEBQ01016134 JAT23843.1 NEVH01013243 PNF29481.1 GGFM01002163 MBW22914.1 GL451538 EFN79080.1 GGFK01004706 MBW38027.1 KK107182 EZA55922.1 PNF29482.1 GECZ01019746 JAS50023.1 LBMM01004381 KMQ92485.1 ADMH02001543 ETN62117.1 KQ434905 KZC11282.1 ATLV01013833 KE524901 KFB38189.1 GL436037 EFN72396.1 GFDL01015375 JAV19670.1 GFDL01015373 JAV19672.1 GGFJ01006531 MBW55672.1 GAMD01002496 JAA99094.1 DS235199 EEB13167.1 KQ971352 EFA06786.1 JXUM01056776 KQ561925 KXJ77137.1 GEDC01031361 GEDC01000561 JAS05937.1 JAS36737.1 CH477265 EAT45572.1 KQ435736 KOX77030.1 KZ288292 PBC29101.1 NNAY01001403 OXU24096.1 GEZM01013390 JAV92600.1 BT127228 AEE62190.1 AJWK01002347 AJWK01002348 AJWK01002349 APGK01034997 KB740923 ENN78130.1 GBYB01007669 JAG77436.1 GANO01002778 JAB57093.1 GDAI01002126 JAI15477.1 GFDF01006612 JAV07472.1 GFDF01006611 JAV07473.1 CM000160 EDW96840.1 AE014297 AY095041 AM294285 AM294287 AM294288 AM294289 AM294290 AM294291 AM294292 AM294293 AM294294 AM294295 FM245353 FM245354 FM245355 FM245356 FM245357 FM245358 FM245360 FM245361 FM245362 FM245363 AAF54556.1 AAM11369.1 CAL26189.1 CAL26191.1 CAL26206.1 CAL26207.1 CAL26208.1 CAL26209.1 CAL26210.1 CAL26211.1 CAL26212.1 CAL26213.1 CAR93279.1 CAR93280.1 CAR93281.1 CAR93282.1 CAR93283.1 CAR93284.1 CAR93286.1 CAR93287.1 CAR93288.1 CAR93289.1 CM000364 EDX13453.1 AM294286 CAL26190.1 CH964272 EDW83984.2 AM294296 CAL26214.1 OUUW01000007 SPP83215.1 CH954181 EDV49600.1 SPP83214.1 CH940652 EDW58908.2 GALA01000244 JAA94608.1 GAMC01000341 JAC06215.1 CH933806 EDW13708.2 FM245359 CAR93285.1 KK120939 KFM79421.1 GAKP01000318 JAC58634.1 GDHF01015385 GDHF01001201 JAI36929.1 JAI51113.1 CM000070 EAL28236.3 CH479182 EDW33838.1 JRES01001539 KNC22170.1 GBXI01000145 JAD14147.1 CH902617 EDV42281.2 CH916369 EDV93864.1 GFWV01019390 MAA44118.1 GFDG01002635 JAV16164.1
SOQ35091.1 HM449859 ADO32994.1 KP735964 AKQ99124.1 KQ459596 KPI95264.1 GAIX01006431 JAA86129.1 AGBW02011765 OWR46171.1 PYGN01000582 PSN43857.1 AAAB01008851 EAA07364.5 APCN01002209 AXCN02000107 GEBQ01012733 JAT27244.1 LJIG01009176 KRT83604.1 KQ978739 KYN28715.1 GL888070 EGI68024.1 KQ981305 KYN42900.1 GECZ01003245 JAS66524.1 ADTU01009939 KK853567 KDR06666.1 DS231863 EDS39975.1 KQ982686 KYQ52295.1 AXCM01004252 QOIP01000011 RLU17174.1 GL767819 EFZ13091.1 GEBQ01016134 JAT23843.1 NEVH01013243 PNF29481.1 GGFM01002163 MBW22914.1 GL451538 EFN79080.1 GGFK01004706 MBW38027.1 KK107182 EZA55922.1 PNF29482.1 GECZ01019746 JAS50023.1 LBMM01004381 KMQ92485.1 ADMH02001543 ETN62117.1 KQ434905 KZC11282.1 ATLV01013833 KE524901 KFB38189.1 GL436037 EFN72396.1 GFDL01015375 JAV19670.1 GFDL01015373 JAV19672.1 GGFJ01006531 MBW55672.1 GAMD01002496 JAA99094.1 DS235199 EEB13167.1 KQ971352 EFA06786.1 JXUM01056776 KQ561925 KXJ77137.1 GEDC01031361 GEDC01000561 JAS05937.1 JAS36737.1 CH477265 EAT45572.1 KQ435736 KOX77030.1 KZ288292 PBC29101.1 NNAY01001403 OXU24096.1 GEZM01013390 JAV92600.1 BT127228 AEE62190.1 AJWK01002347 AJWK01002348 AJWK01002349 APGK01034997 KB740923 ENN78130.1 GBYB01007669 JAG77436.1 GANO01002778 JAB57093.1 GDAI01002126 JAI15477.1 GFDF01006612 JAV07472.1 GFDF01006611 JAV07473.1 CM000160 EDW96840.1 AE014297 AY095041 AM294285 AM294287 AM294288 AM294289 AM294290 AM294291 AM294292 AM294293 AM294294 AM294295 FM245353 FM245354 FM245355 FM245356 FM245357 FM245358 FM245360 FM245361 FM245362 FM245363 AAF54556.1 AAM11369.1 CAL26189.1 CAL26191.1 CAL26206.1 CAL26207.1 CAL26208.1 CAL26209.1 CAL26210.1 CAL26211.1 CAL26212.1 CAL26213.1 CAR93279.1 CAR93280.1 CAR93281.1 CAR93282.1 CAR93283.1 CAR93284.1 CAR93286.1 CAR93287.1 CAR93288.1 CAR93289.1 CM000364 EDX13453.1 AM294286 CAL26190.1 CH964272 EDW83984.2 AM294296 CAL26214.1 OUUW01000007 SPP83215.1 CH954181 EDV49600.1 SPP83214.1 CH940652 EDW58908.2 GALA01000244 JAA94608.1 GAMC01000341 JAC06215.1 CH933806 EDW13708.2 FM245359 CAR93285.1 KK120939 KFM79421.1 GAKP01000318 JAC58634.1 GDHF01015385 GDHF01001201 JAI36929.1 JAI51113.1 CM000070 EAL28236.3 CH479182 EDW33838.1 JRES01001539 KNC22170.1 GBXI01000145 JAD14147.1 CH902617 EDV42281.2 CH916369 EDV93864.1 GFWV01019390 MAA44118.1 GFDG01002635 JAV16164.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000245037
+ More
UP000075903 UP000075882 UP000075902 UP000076407 UP000007062 UP000075840 UP000075885 UP000075884 UP000075886 UP000075900 UP000076408 UP000075901 UP000078492 UP000007755 UP000078541 UP000005205 UP000027135 UP000002320 UP000075809 UP000075883 UP000075920 UP000279307 UP000235965 UP000008237 UP000053097 UP000036403 UP000075881 UP000000673 UP000076502 UP000030765 UP000000311 UP000009046 UP000007266 UP000075880 UP000069940 UP000249989 UP000008820 UP000192223 UP000053105 UP000242457 UP000215335 UP000092461 UP000019118 UP000002282 UP000000803 UP000000304 UP000192221 UP000007798 UP000268350 UP000008711 UP000008792 UP000009192 UP000054359 UP000001819 UP000008744 UP000037069 UP000007801 UP000001070
UP000075903 UP000075882 UP000075902 UP000076407 UP000007062 UP000075840 UP000075885 UP000075884 UP000075886 UP000075900 UP000076408 UP000075901 UP000078492 UP000007755 UP000078541 UP000005205 UP000027135 UP000002320 UP000075809 UP000075883 UP000075920 UP000279307 UP000235965 UP000008237 UP000053097 UP000036403 UP000075881 UP000000673 UP000076502 UP000030765 UP000000311 UP000009046 UP000007266 UP000075880 UP000069940 UP000249989 UP000008820 UP000192223 UP000053105 UP000242457 UP000215335 UP000092461 UP000019118 UP000002282 UP000000803 UP000000304 UP000192221 UP000007798 UP000268350 UP000008711 UP000008792 UP000009192 UP000054359 UP000001819 UP000008744 UP000037069 UP000007801 UP000001070
Pfam
PF13649 Methyltransf_25
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J5I6
A0A2A4JZ74
A0A3S2P291
A0A2H1V2N3
E3UKI5
A0A0K0PYY8
+ More
A0A194PR89 S4P7D1 A0A212EXI1 A0A2P8YHX0 A0A182V6T4 A0A182KX41 A0A182TPM7 A0A182XK30 Q7PRE6 A0A182HJW1 A0A182PN09 A0A182NKY4 A0A182QNV1 A0A1B6LU91 A0A0T6B8E5 A0A182R846 A0A182YM54 A0A182T1Q6 A0A195EKC3 F4WC44 A0A195FQP0 A0A1B6GVP5 A0A158NA18 A0A067QQH5 B0W9B5 A0A151WXA2 A0A182M8C9 A0A182VS92 A0A3L8DA14 E9J265 A0A1B6LJG4 A0A2J7QLL8 A0A2M3Z313 E2BZ08 A0A2M4AB66 A0A026WIQ2 A0A2J7QLM3 A0A1B6FJ66 A0A0J7NIZ7 A0A182K728 W5JCN8 A0A154PHI8 A0A084VJP5 E2A2D3 A0A1Q3EWJ4 A0A1Q3EWI3 A0A2M4BRM9 T1DH59 E0VIG1 D6WU94 A0A182IWC5 A0A182GDW4 A0A1B6EFM7 Q17G76 A0A1S4F430 A0A1W4WQZ3 A0A0M9A4F1 A0A2A3EDK7 A0A232F014 A0A1Y1N6P3 J3JVG0 A0A1B0C9H2 N6U8X7 A0A0C9QVA8 U5ETH4 A0A0K8TM86 A0A1L8DLZ2 A0A1L8DLX2 B4PLD3 Q9VGW7 B4QUP6 A0A1W4W0T7 A0ANP8 B4NJY2 A0ANQ8 A0A3B0JLV3 B3P1K1 A0A3B0JLL6 B4M5I2 T1E306 W8CEE4 B4KC99 C0MJ37 A0A087UPY2 A0A034WVH2 A0A0K8WIY1 Q297G9 B4GE03 A0A0L0BQ09 A0A0A1XSZ8 B3M290 B4JHM5 A0A293MW14 A0A1L8EBY0
A0A194PR89 S4P7D1 A0A212EXI1 A0A2P8YHX0 A0A182V6T4 A0A182KX41 A0A182TPM7 A0A182XK30 Q7PRE6 A0A182HJW1 A0A182PN09 A0A182NKY4 A0A182QNV1 A0A1B6LU91 A0A0T6B8E5 A0A182R846 A0A182YM54 A0A182T1Q6 A0A195EKC3 F4WC44 A0A195FQP0 A0A1B6GVP5 A0A158NA18 A0A067QQH5 B0W9B5 A0A151WXA2 A0A182M8C9 A0A182VS92 A0A3L8DA14 E9J265 A0A1B6LJG4 A0A2J7QLL8 A0A2M3Z313 E2BZ08 A0A2M4AB66 A0A026WIQ2 A0A2J7QLM3 A0A1B6FJ66 A0A0J7NIZ7 A0A182K728 W5JCN8 A0A154PHI8 A0A084VJP5 E2A2D3 A0A1Q3EWJ4 A0A1Q3EWI3 A0A2M4BRM9 T1DH59 E0VIG1 D6WU94 A0A182IWC5 A0A182GDW4 A0A1B6EFM7 Q17G76 A0A1S4F430 A0A1W4WQZ3 A0A0M9A4F1 A0A2A3EDK7 A0A232F014 A0A1Y1N6P3 J3JVG0 A0A1B0C9H2 N6U8X7 A0A0C9QVA8 U5ETH4 A0A0K8TM86 A0A1L8DLZ2 A0A1L8DLX2 B4PLD3 Q9VGW7 B4QUP6 A0A1W4W0T7 A0ANP8 B4NJY2 A0ANQ8 A0A3B0JLV3 B3P1K1 A0A3B0JLL6 B4M5I2 T1E306 W8CEE4 B4KC99 C0MJ37 A0A087UPY2 A0A034WVH2 A0A0K8WIY1 Q297G9 B4GE03 A0A0L0BQ09 A0A0A1XSZ8 B3M290 B4JHM5 A0A293MW14 A0A1L8EBY0
PDB
4X41
E-value=3.58574e-140,
Score=1276
Ontologies
GO
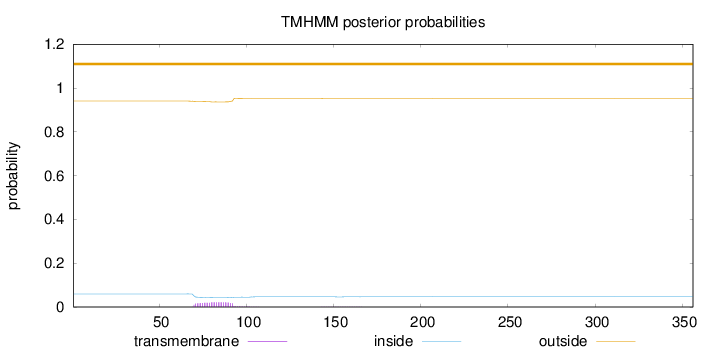
Topology
Length:
356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48262
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05976
outside
1 - 356
Population Genetic Test Statistics
Pi
257.661207
Theta
207.235191
Tajima's D
0.3141
CLR
0.406006
CSRT
0.458327083645818
Interpretation
Uncertain
