Gene
KWMTBOMO15409
Annotation
PREDICTED:_probable_cleavage_and_polyadenylation_specificity_factor_subunit_2?_partial_[Bombyx_mori]
Full name
Cleavage and polyadenylation specificity factor subunit 2
+ More
Probable cleavage and polyadenylation specificity factor subunit 2
Probable cleavage and polyadenylation specificity factor subunit 2
Alternative Name
Cleavage and polyadenylation specificity factor 100 kDa subunit
Location in the cell
Cytoplasmic Reliability : 3.038
Sequence
CDS
ATGACTTCAATAATTAAATTACATTGCTTATCTGGTGCTGGTGAGGAAACTCCTCCGTGCTATGTGCTCCAAATAGATGAATTCAAATTCCTCCTGGATTGCGGATGGGACGAGAAATTCGATATGGTTTTCATTAAAGAACTTAAAAGACATGTAAATTCAATAGATGCTGTACTAATATCACACTCGGATCCTCTACATTTAGGAGCTCTTCCATATGCAGTTGGAAAACTCGGTCTTAGCTGTCCAATATATGCAACTCTACCGGTATACAAAATGGGGCAGATGTTCATGTACGATTTATACCAAGCCCATAAGAATGTGTCCGAGTTCGATTTGTTCACACTGGATGATGTTGATATGGCTTTTGACAGAATCGTACAGTTGAAATACAATCAGAGCATTGAAATGAAAGGAAAAGGTCTAGGTCTCCGCATTACACCACTCCCAGCTGGCCATTCCCTTGGTGGAACAATATGGCGCATATCCGCCCCCGGCGAGGAAGATATTGTTTACGCGCCTGACTTCAACCACAAGAAAGAGAGACATCTGAATGGATGTGACATCGAAAGACTGATGAGACCCTCTCTGATGCTGTTGGGAGCTATGAATGCTGATTACGTGCAACAGAGGAGAAGACTGAGAGATGAAAAATTGATGACAACAATTCTTACAACGCTGCGAGGAGGCGGTTGCGTTTTGGTATGCACAGACACAGCCGGTCGAGTCCTAGAATTGGCCCACATGCTGGACCAACTGTGGAGGAACAAGGATTCCGGGCTCGTGGCCTATTCGTTACTTCTCCTGTCCAACGTCAGTTACAATGTCGTAGAGTTCGCTAAATCTCAGATAGAGTGGATGAGCGACAAACTGACGAGGGCGTTTGAAGGGGCTCGCAGTAATCCCTTTGCTTTGAGACACGTGCAGCTGTGTCACTCGGCCGGTGACGTCACTAGGACCCCGGGACCGAAGGTCGTCCTGGCCTCGTTCCCGGACCTGGAGGCGGGCTTCGCCAGAGACCTGTTCCTCCAGTGGGCCCCACACTCCCACAACTCGATCGTGCTGACGGCCAGAACATCCCCCGGCACACTGGCAAGAGATCTGATAGACAACGGGGGGGACCGCGTCATCGAGCTGGTGGTTCGACGGAGGGTGAAGCTGGAGGGCGCCGAGCTCGAGGAGTTCCTGCAGCAGCAGAGAATGAACGTCACCAATACCGTGAGGGAGGAGGTGGCCGGCATCTCGTCGGACTCTGACTCCGAAGACGAGCTGGAGATGTGCGTGGTGACCGGGAGACACGACATCCCCGTGCGCCTGGAGACCAGGCTGGGCGGCTGCTTCAAGTCCAACAAGCGGCACCACGCCATCTACCCCTTCCACGAGGAGCGGACCAGGGCTGATGACTACGGAGAGATCATTAAGCCAGAAGATTACAGACTTGCCGAGATAGTGGACGCCGAAGGGGAGATCCGGGACGTGCCCCCCACTGCCCCCATCAAACAGGAACAAGATGAGGAAATGTCGGAGGTCCCGAGCAAGTGCGTGTCCTCGGTGCGGCAGGTACACGTCAAGGCGCACGTCGAGTACATCGAGCTGGAAGGGCGCTGCGATGGGGAGTCCCTGCTCAGGGTCGTGTCGCAGACCAAGCCCTGGGCAGTGGTGGGGCTCCGGGCCAGCGAGGCGGCGCTGCACACCCTCAAAAAACATTGTGAGTCTGAAGGAATCGAAAAGGTGTTCTTACCGTCCGTTGGGGAAACCGTCGACGCCACGACAGAGTCACACATCTATCAGGTGAAACTGACGGACGGGCTGATGCGGGGCCTGGTGTGGCGGCAGGCCGGGGACGCCGAGCTGGCCTGGCTGTCCGCCGTCGTGGCGCCGCGCACGCACGCCCGGGAGAGCGACGCCGCCCCCGAGGAGGGCGGCGGTGCGGTGTCGCTGGAGGCGGGTGCGGGGCGGCCGCGCGCGGCTGCGTTCGTGAACACGGTGCGGCTGCACGAGCTGCGCGCCGCCCTCGCCCGCGCCGGCCTGCAGCCCGACCTCGGCGCCGGCACGCTCACCTGCTGCTCCGGGACGCTCGCCGTGCGCAGGATGGAAAACGGCCAAGTTTCTTTAGAAGGAGTCCTGTCTGAAGAGTATTACAAAGTGCGAGAACTCTTGTACGAACAATTCGCGATAGTCTGA
Protein
MTSIIKLHCLSGAGEETPPCYVLQIDEFKFLLDCGWDEKFDMVFIKELKRHVNSIDAVLISHSDPLHLGALPYAVGKLGLSCPIYATLPVYKMGQMFMYDLYQAHKNVSEFDLFTLDDVDMAFDRIVQLKYNQSIEMKGKGLGLRITPLPAGHSLGGTIWRISAPGEEDIVYAPDFNHKKERHLNGCDIERLMRPSLMLLGAMNADYVQQRRRLRDEKLMTTILTTLRGGGCVLVCTDTAGRVLELAHMLDQLWRNKDSGLVAYSLLLLSNVSYNVVEFAKSQIEWMSDKLTRAFEGARSNPFALRHVQLCHSAGDVTRTPGPKVVLASFPDLEAGFARDLFLQWAPHSHNSIVLTARTSPGTLARDLIDNGGDRVIELVVRRRVKLEGAELEEFLQQQRMNVTNTVREEVAGISSDSDSEDELEMCVVTGRHDIPVRLETRLGGCFKSNKRHHAIYPFHEERTRADDYGEIIKPEDYRLAEIVDAEGEIRDVPPTAPIKQEQDEEMSEVPSKCVSSVRQVHVKAHVEYIELEGRCDGESLLRVVSQTKPWAVVGLRASEAALHTLKKHCESEGIEKVFLPSVGETVDATTESHIYQVKLTDGLMRGLVWRQAGDAELAWLSAVVAPRTHARESDAAPEEGGGAVSLEAGAGRPRAAAFVNTVRLHELRAALARAGLQPDLGAGTLTCCSGTLAVRRMENGQVSLEGVLSEEYYKVRELLYEQFAIV
Summary
Description
Component of the cleavage and polyadenylation specificity factor (CPSF) complex that plays a key role in pre-mRNA 3'-end formation, recognizing the AAUAAA signal sequence and interacting with poly(A) polymerase and other factors to bring about cleavage and poly(A) addition. Required for the cotranscriptional processing of 3'-ends of polyadenylated and histone pre-mRNA.
Subunit
Component of the cleavage and polyadenylation specificity factor (CPSF) complex, composed of at least Clp, Cpsf73, Cpsf100 and Cpsf160. Interacts with Sym and Cpsf73 forming a core cleavage factor required for both polyadenylated and histone mRNA processing. Interacts with Slbp and Lsm11.
Similarity
Belongs to the metallo-beta-lactamase superfamily. RNA-metabolizing metallo-beta-lactamase-like family. CPSF2/YSH1 subfamily.
Keywords
Complete proteome
mRNA processing
Nucleus
Phosphoprotein
Reference proteome
RNA-binding
Feature
chain Probable cleavage and polyadenylation specificity factor subunit 2
Uniprot
A0A2A4JZW4
S4P5K0
A0A194Q7M5
A0A212FPY6
D6WP71
A0A1W4WPS5
+ More
A0A067RD17 A0A1I8NZS9 A0A1B6EDG6 E9J8R8 F4WAT5 A0A151IHA7 A0A151IV75 A0A0K8UAC7 A0A158P3K2 K7J1K2 A0A195F093 A0A3L8DH36 A0A151XDK9 A0A232EY66 E1ZX64 A0A154PH25 A0A026WYH7 A0A0A1X2P0 A0A2A3EIW9 A0A1Y1M0V5 A0A087ZQ62 Q17Q35 A0A310SIA7 W8BAF9 A0A0L7R440 A0A2J7QXR1 A0A1I8M3S6 A0A3B0KHW8 A0A182H7P4 A0A1S4EUZ6 A0A034WQU1 A0A182FT51 W5JIQ1 A0A0P6IU15 A0A1B0CL15 A0A195BYZ2 A0A1A9X1U9 A0A1B0GEA0 A0A1A9US68 B4PQ87 B3P5M6 A0A0C9QTZ5 Q9V3D6 N6T7L6 A0A336LAG3 B4JTB6 A0A1W4VVQ4 E0VYY6 B3LXN9 A0A182Q328 A0A182JRJ0 A0A2M4BEN8 A0A182R368 B4KDT1 A0A182VXE7 Q7QC68 B4HZ45 B0WQG5 A0A084WRC4 A0A182M9G5 B4NF98 A0A182XWV9 A0A182T530 A0A182PAF6 A0A182XI04 A0A182LF29 A0A182HSY5 A0A182IJ44 A0A182VCI4 A0A182TFD9 A0A1A9YBU3 A0A0M4F298 A0A1B0BXC6 A0A182NLA6 T1J0U4 A0A1J1I7W1 B5DYG6 A0A0A9YR18 A0A0K8SNA9 A0A2R7WYN0 A0A1Q3F2G1 A0A1S3HFG1 A0A0V0G937 A0A023F389 R7V1Z3 A0A131YJH7 A0A0M9A4Q2 A0A1A9Z6W8 A0A0P4VNT6 A0A1E1XLS1 E9H4K8 A0A087SZA3 A0A3Q3B974
A0A067RD17 A0A1I8NZS9 A0A1B6EDG6 E9J8R8 F4WAT5 A0A151IHA7 A0A151IV75 A0A0K8UAC7 A0A158P3K2 K7J1K2 A0A195F093 A0A3L8DH36 A0A151XDK9 A0A232EY66 E1ZX64 A0A154PH25 A0A026WYH7 A0A0A1X2P0 A0A2A3EIW9 A0A1Y1M0V5 A0A087ZQ62 Q17Q35 A0A310SIA7 W8BAF9 A0A0L7R440 A0A2J7QXR1 A0A1I8M3S6 A0A3B0KHW8 A0A182H7P4 A0A1S4EUZ6 A0A034WQU1 A0A182FT51 W5JIQ1 A0A0P6IU15 A0A1B0CL15 A0A195BYZ2 A0A1A9X1U9 A0A1B0GEA0 A0A1A9US68 B4PQ87 B3P5M6 A0A0C9QTZ5 Q9V3D6 N6T7L6 A0A336LAG3 B4JTB6 A0A1W4VVQ4 E0VYY6 B3LXN9 A0A182Q328 A0A182JRJ0 A0A2M4BEN8 A0A182R368 B4KDT1 A0A182VXE7 Q7QC68 B4HZ45 B0WQG5 A0A084WRC4 A0A182M9G5 B4NF98 A0A182XWV9 A0A182T530 A0A182PAF6 A0A182XI04 A0A182LF29 A0A182HSY5 A0A182IJ44 A0A182VCI4 A0A182TFD9 A0A1A9YBU3 A0A0M4F298 A0A1B0BXC6 A0A182NLA6 T1J0U4 A0A1J1I7W1 B5DYG6 A0A0A9YR18 A0A0K8SNA9 A0A2R7WYN0 A0A1Q3F2G1 A0A1S3HFG1 A0A0V0G937 A0A023F389 R7V1Z3 A0A131YJH7 A0A0M9A4Q2 A0A1A9Z6W8 A0A0P4VNT6 A0A1E1XLS1 E9H4K8 A0A087SZA3 A0A3Q3B974
Pubmed
23622113
26354079
22118469
18362917
19820115
24845553
+ More
21282665 21719571 21347285 20075255 30249741 28648823 20798317 24508170 25830018 28004739 17510324 24495485 25315136 26483478 25348373 20920257 23761445 26999592 17994087 17550304 10731132 12537572 10731138 18327897 19450530 23537049 20566863 12364791 24438588 25244985 20966253 15632085 25401762 25474469 23254933 26830274 27129103 29209593 21292972
21282665 21719571 21347285 20075255 30249741 28648823 20798317 24508170 25830018 28004739 17510324 24495485 25315136 26483478 25348373 20920257 23761445 26999592 17994087 17550304 10731132 12537572 10731138 18327897 19450530 23537049 20566863 12364791 24438588 25244985 20966253 15632085 25401762 25474469 23254933 26830274 27129103 29209593 21292972
EMBL
NWSH01000346
PCG77218.1
GAIX01005494
JAA87066.1
KQ459324
KPJ01523.1
+ More
AGBW02000323 OWR55794.1 KQ971352 EFA07272.1 KK852540 KDR21766.1 GEDC01001336 JAS35962.1 GL769036 EFZ10838.1 GL888053 EGI68691.1 KQ977636 KYN01143.1 KQ980924 KYN11412.1 GDHF01028682 JAI23632.1 ADTU01008167 KQ981880 KYN33898.1 QOIP01000008 RLU19492.1 KQ982268 KYQ58464.1 NNAY01001671 OXU23263.1 GL435030 EFN74154.1 KQ434904 KZC11186.1 KK107064 EZA60898.1 GBXI01009364 JAD04928.1 KZ288229 PBC31715.1 GEZM01044356 JAV78280.1 CH477187 EAT48881.1 KQ767768 OAD53310.1 GAMC01012537 JAB94018.1 KQ414658 KOC65642.1 NEVH01009372 PNF33368.1 OUUW01000013 SPP88060.1 JXUM01116783 KQ566020 KXJ70452.1 GAKP01002834 JAC56118.1 ADMH02001133 ETN63991.1 GDUN01000593 JAN95326.1 AJWK01016831 KQ976394 KYM93138.1 CCAG010012178 CM000160 EDW98356.2 CH954182 EDV53276.1 GBYB01007214 GBYB01012073 GBYB01012076 GBYB01012077 JAG76981.1 JAG81840.1 JAG81843.1 JAG81844.1 AE014297 AF160933 APGK01049397 KB741150 KB632095 ENN73673.1 ERL88780.1 UFQS01001426 UFQT01001426 SSX10828.1 SSX30508.1 CH916373 EDV95006.1 DS235848 EEB18592.1 CH902617 EDV43933.1 AXCN02000116 GGFJ01002374 MBW51515.1 CH933806 EDW14928.1 AAAB01008859 EAA08192.4 CH480819 EDW53302.1 DS232039 EDS32836.1 ATLV01026020 KE525405 KFB52768.1 AXCM01000492 CH964251 EDW82965.1 APCN01000376 CP012526 ALC45525.1 JXJN01022185 AFFK01020674 CVRI01000043 CRK96399.1 CM000070 EDY67988.3 GBHO01009538 JAG34066.1 GBRD01011091 JAG54733.1 KK856018 PTY24658.1 GFDL01013302 JAV21743.1 GECL01001727 JAP04397.1 GBBI01002994 JAC15718.1 AMQN01000769 KB295623 ELU12868.1 GEDV01009440 JAP79117.1 KQ435756 KOX75893.1 GDKW01000222 JAI56373.1 GFAA01003493 JAT99941.1 GL732592 EFX73157.1 KK112648 KFM58192.1
AGBW02000323 OWR55794.1 KQ971352 EFA07272.1 KK852540 KDR21766.1 GEDC01001336 JAS35962.1 GL769036 EFZ10838.1 GL888053 EGI68691.1 KQ977636 KYN01143.1 KQ980924 KYN11412.1 GDHF01028682 JAI23632.1 ADTU01008167 KQ981880 KYN33898.1 QOIP01000008 RLU19492.1 KQ982268 KYQ58464.1 NNAY01001671 OXU23263.1 GL435030 EFN74154.1 KQ434904 KZC11186.1 KK107064 EZA60898.1 GBXI01009364 JAD04928.1 KZ288229 PBC31715.1 GEZM01044356 JAV78280.1 CH477187 EAT48881.1 KQ767768 OAD53310.1 GAMC01012537 JAB94018.1 KQ414658 KOC65642.1 NEVH01009372 PNF33368.1 OUUW01000013 SPP88060.1 JXUM01116783 KQ566020 KXJ70452.1 GAKP01002834 JAC56118.1 ADMH02001133 ETN63991.1 GDUN01000593 JAN95326.1 AJWK01016831 KQ976394 KYM93138.1 CCAG010012178 CM000160 EDW98356.2 CH954182 EDV53276.1 GBYB01007214 GBYB01012073 GBYB01012076 GBYB01012077 JAG76981.1 JAG81840.1 JAG81843.1 JAG81844.1 AE014297 AF160933 APGK01049397 KB741150 KB632095 ENN73673.1 ERL88780.1 UFQS01001426 UFQT01001426 SSX10828.1 SSX30508.1 CH916373 EDV95006.1 DS235848 EEB18592.1 CH902617 EDV43933.1 AXCN02000116 GGFJ01002374 MBW51515.1 CH933806 EDW14928.1 AAAB01008859 EAA08192.4 CH480819 EDW53302.1 DS232039 EDS32836.1 ATLV01026020 KE525405 KFB52768.1 AXCM01000492 CH964251 EDW82965.1 APCN01000376 CP012526 ALC45525.1 JXJN01022185 AFFK01020674 CVRI01000043 CRK96399.1 CM000070 EDY67988.3 GBHO01009538 JAG34066.1 GBRD01011091 JAG54733.1 KK856018 PTY24658.1 GFDL01013302 JAV21743.1 GECL01001727 JAP04397.1 GBBI01002994 JAC15718.1 AMQN01000769 KB295623 ELU12868.1 GEDV01009440 JAP79117.1 KQ435756 KOX75893.1 GDKW01000222 JAI56373.1 GFAA01003493 JAT99941.1 GL732592 EFX73157.1 KK112648 KFM58192.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000007266
UP000192223
UP000027135
+ More
UP000095300 UP000007755 UP000078542 UP000078492 UP000005205 UP000002358 UP000078541 UP000279307 UP000075809 UP000215335 UP000000311 UP000076502 UP000053097 UP000242457 UP000005203 UP000008820 UP000053825 UP000235965 UP000095301 UP000268350 UP000069940 UP000249989 UP000069272 UP000000673 UP000092461 UP000078540 UP000091820 UP000092444 UP000078200 UP000002282 UP000008711 UP000000803 UP000019118 UP000030742 UP000001070 UP000192221 UP000009046 UP000007801 UP000075886 UP000075881 UP000075900 UP000009192 UP000075920 UP000007062 UP000001292 UP000002320 UP000030765 UP000075883 UP000007798 UP000076408 UP000075901 UP000075885 UP000076407 UP000075882 UP000075840 UP000075880 UP000075903 UP000075902 UP000092443 UP000092553 UP000092460 UP000075884 UP000183832 UP000001819 UP000085678 UP000014760 UP000053105 UP000092445 UP000000305 UP000054359 UP000264800
UP000095300 UP000007755 UP000078542 UP000078492 UP000005205 UP000002358 UP000078541 UP000279307 UP000075809 UP000215335 UP000000311 UP000076502 UP000053097 UP000242457 UP000005203 UP000008820 UP000053825 UP000235965 UP000095301 UP000268350 UP000069940 UP000249989 UP000069272 UP000000673 UP000092461 UP000078540 UP000091820 UP000092444 UP000078200 UP000002282 UP000008711 UP000000803 UP000019118 UP000030742 UP000001070 UP000192221 UP000009046 UP000007801 UP000075886 UP000075881 UP000075900 UP000009192 UP000075920 UP000007062 UP000001292 UP000002320 UP000030765 UP000075883 UP000007798 UP000076408 UP000075901 UP000075885 UP000076407 UP000075882 UP000075840 UP000075880 UP000075903 UP000075902 UP000092443 UP000092553 UP000092460 UP000075884 UP000183832 UP000001819 UP000085678 UP000014760 UP000053105 UP000092445 UP000000305 UP000054359 UP000264800
PRIDE
Interpro
SUPFAM
SSF56281
SSF56281
Gene 3D
ProteinModelPortal
A0A2A4JZW4
S4P5K0
A0A194Q7M5
A0A212FPY6
D6WP71
A0A1W4WPS5
+ More
A0A067RD17 A0A1I8NZS9 A0A1B6EDG6 E9J8R8 F4WAT5 A0A151IHA7 A0A151IV75 A0A0K8UAC7 A0A158P3K2 K7J1K2 A0A195F093 A0A3L8DH36 A0A151XDK9 A0A232EY66 E1ZX64 A0A154PH25 A0A026WYH7 A0A0A1X2P0 A0A2A3EIW9 A0A1Y1M0V5 A0A087ZQ62 Q17Q35 A0A310SIA7 W8BAF9 A0A0L7R440 A0A2J7QXR1 A0A1I8M3S6 A0A3B0KHW8 A0A182H7P4 A0A1S4EUZ6 A0A034WQU1 A0A182FT51 W5JIQ1 A0A0P6IU15 A0A1B0CL15 A0A195BYZ2 A0A1A9X1U9 A0A1B0GEA0 A0A1A9US68 B4PQ87 B3P5M6 A0A0C9QTZ5 Q9V3D6 N6T7L6 A0A336LAG3 B4JTB6 A0A1W4VVQ4 E0VYY6 B3LXN9 A0A182Q328 A0A182JRJ0 A0A2M4BEN8 A0A182R368 B4KDT1 A0A182VXE7 Q7QC68 B4HZ45 B0WQG5 A0A084WRC4 A0A182M9G5 B4NF98 A0A182XWV9 A0A182T530 A0A182PAF6 A0A182XI04 A0A182LF29 A0A182HSY5 A0A182IJ44 A0A182VCI4 A0A182TFD9 A0A1A9YBU3 A0A0M4F298 A0A1B0BXC6 A0A182NLA6 T1J0U4 A0A1J1I7W1 B5DYG6 A0A0A9YR18 A0A0K8SNA9 A0A2R7WYN0 A0A1Q3F2G1 A0A1S3HFG1 A0A0V0G937 A0A023F389 R7V1Z3 A0A131YJH7 A0A0M9A4Q2 A0A1A9Z6W8 A0A0P4VNT6 A0A1E1XLS1 E9H4K8 A0A087SZA3 A0A3Q3B974
A0A067RD17 A0A1I8NZS9 A0A1B6EDG6 E9J8R8 F4WAT5 A0A151IHA7 A0A151IV75 A0A0K8UAC7 A0A158P3K2 K7J1K2 A0A195F093 A0A3L8DH36 A0A151XDK9 A0A232EY66 E1ZX64 A0A154PH25 A0A026WYH7 A0A0A1X2P0 A0A2A3EIW9 A0A1Y1M0V5 A0A087ZQ62 Q17Q35 A0A310SIA7 W8BAF9 A0A0L7R440 A0A2J7QXR1 A0A1I8M3S6 A0A3B0KHW8 A0A182H7P4 A0A1S4EUZ6 A0A034WQU1 A0A182FT51 W5JIQ1 A0A0P6IU15 A0A1B0CL15 A0A195BYZ2 A0A1A9X1U9 A0A1B0GEA0 A0A1A9US68 B4PQ87 B3P5M6 A0A0C9QTZ5 Q9V3D6 N6T7L6 A0A336LAG3 B4JTB6 A0A1W4VVQ4 E0VYY6 B3LXN9 A0A182Q328 A0A182JRJ0 A0A2M4BEN8 A0A182R368 B4KDT1 A0A182VXE7 Q7QC68 B4HZ45 B0WQG5 A0A084WRC4 A0A182M9G5 B4NF98 A0A182XWV9 A0A182T530 A0A182PAF6 A0A182XI04 A0A182LF29 A0A182HSY5 A0A182IJ44 A0A182VCI4 A0A182TFD9 A0A1A9YBU3 A0A0M4F298 A0A1B0BXC6 A0A182NLA6 T1J0U4 A0A1J1I7W1 B5DYG6 A0A0A9YR18 A0A0K8SNA9 A0A2R7WYN0 A0A1Q3F2G1 A0A1S3HFG1 A0A0V0G937 A0A023F389 R7V1Z3 A0A131YJH7 A0A0M9A4Q2 A0A1A9Z6W8 A0A0P4VNT6 A0A1E1XLS1 E9H4K8 A0A087SZA3 A0A3Q3B974
PDB
6I1D
E-value=1.48599e-27,
Score=308
Ontologies
GO
PANTHER
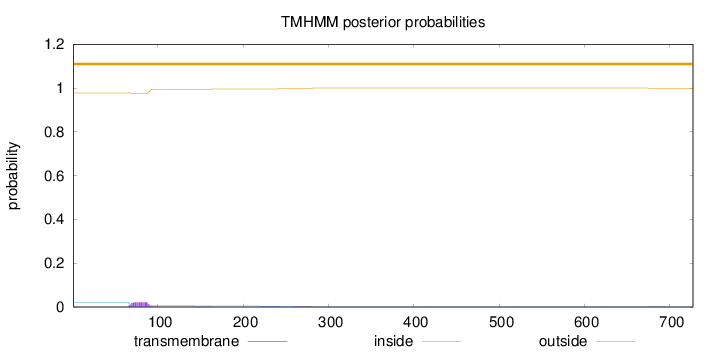
Topology
Subcellular location
Nucleus
Length:
727
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.603359999999999
Exp number, first 60 AAs:
0.00066
Total prob of N-in:
0.02211
outside
1 - 727
Population Genetic Test Statistics
Pi
265.834437
Theta
167.892463
Tajima's D
1.923741
CLR
0.361363
CSRT
0.872056397180141
Interpretation
Uncertain
