Pre Gene Modal
BGIBMGA004891
Annotation
transmembrane_trafficking_protein_precursor_[Bombyx_mori]
Full name
Transmembrane emp24 domain-containing protein bai
Alternative Name
Protein baiser
Location in the cell
Cytoplasmic Reliability : 1.604 Extracellular Reliability : 1.827
Sequence
CDS
ATGGAGGTACAATATCTCGTGTTGTTACTGAGCTTAATCTGGCATGGAACTGATGCGATTATGTGGAGCTTGGCGCCGAACACACACAAGTGTTTGAAAGAAGAATTACACGCCAACGTGTTAGTTGCAGGAGAATATGATGTGACAGAAATACAGGGGCAAAGAGTTGACTATATTATTAAAGACTCGAAGGGGCACATCTTATCACAGAAAGATACAGTAACAAAGGGAAAGTTCTCATTCGTCACAGAGAATTATGATATGTTTGAAGTTTGCTTTATATCTAAAGTACCATCAGAAAGAAGAGGTATTCCACATCAAGTAAGCTTAGATATTAAAATCGGCATAGAAGCAAAGACTTATGAAGGGATCGGGGAGGCTGCAAAATTGAAGCCAATGGAAGTGGAACTGAAGAGACTAGAAGACCTATCAGAAGCTATCGTTCAAGATTTCACACTAATGAGGAAAAGAGAAGAAGAAATGAGAGATACCAATGAATCAACGAATAATAGAGTTTTATTCTTCAGTATATTCTCGATGGCGTGCCTCTTGGGTCTGGCCACCTGGCAAGTCCTGTACCTTCGCAGGTTCTTCAAAGCAAAGAAGCTAATTGAGTAA
Protein
MEVQYLVLLLSLIWHGTDAIMWSLAPNTHKCLKEELHANVLVAGEYDVTEIQGQRVDYIIKDSKGHILSQKDTVTKGKFSFVTENYDMFEVCFISKVPSERRGIPHQVSLDIKIGIEAKTYEGIGEAAKLKPMEVELKRLEDLSEAIVQDFTLMRKREEEMRDTNESTNNRVLFFSIFSMACLLGLATWQVLYLRRFFKAKKLIE
Summary
Description
Eca and bai are essential, though not redundant, for dorsoventral patterning of the embryo. Specifically required during early embryogenesis for the activity of maternal tkv, while the zygotic tkv is not affected (By similarity).
Eca and bai are essential, though not redundant, for dorsoventral patterning of the embryo. Specifically required during early embryogenesis for the activity of maternal tkv, while the zygotic tkv is not affected.
Eca and bai are essential, though not redundant, for dorsoventral patterning of the embryo. Specifically required during early embryogenesis for the activity of maternal tkv, while the zygotic tkv is not affected.
Similarity
Belongs to the EMP24/GP25L family.
Keywords
Complete proteome
Developmental protein
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Transmembrane emp24 domain-containing protein bai
Uniprot
Q1HQA9
A0A2W1BJG3
A0A2A4K0C5
A0A2H1WYB3
A0A3G1T1A6
A0A1E1VY15
+ More
A0A3S2LJJ0 I4DKY2 S4PAJ4 A0A0N1I5L7 A0A212FPU6 I4DK59 A0A2P8ZAF1 C4PLE9 A0A1W4XI10 Q7Q2F1 A0A2J7R6M8 Q1HQX4 A0A023EJA5 A0A023ELM8 A0A182RT68 A0A084VQF3 A0A182M718 A0A182VRF2 W5JX87 T1E2D4 A0A2M4A6F9 B3LVB0 B4GE47 A0A2M3Z171 U5EYG5 A0A1Q3EUC2 A0A1Q3EUB7 A0A1Y1KLI0 A0A2M3Z1C6 A0A182FJN9 B4PUZ3 B3P6T8 A0A1W4V8F2 A0A3B0JQK5 A0A2M4BZZ3 T1DL52 Q8SXY6 A0AQ71 A0A182IUG3 A0A1S4EL48 A0A1Q3EUD8 A0A1Q3EUC9 B4K4G5 A0A1A9W6N6 A0A336MYX4 E0VJ28 B4NIY1 B4MGF8 A0A1I8NM88 A0A1I8M6I8 A0A1A9XN09 A0A232EHB8 K7IUJ7 A0A1L8EF07 A0A1B0A8S4 A0A1B0CSV2 A0A1B0DFB6 A0A1L8DUJ1 B4JYU5 R4WQW2 A0A151WJ83 A0A195BG79 A0A195EFI6 A0A195F0Q7 F4WG56 E9IBD7 A0A195C8M4 D7GXL6 A0A1B6D7M5 A0A026WR71 A0A158NT36 A0A1B6MAZ9 A0A1J1I428 A0A0P4W5G7 E2A849 V5GN59 E2BA16 A0A3R7M9P9 A0A0J7LA60 R4UND3 A0A3L8DFM6 E9G6B6 A0A162PFE3 A0A2I9LPT8 A0A1S3II12 A0A2P2HX57 A0A1W7RAJ4 A0A0K8R831 B7P427 A0A1D2N171 C4WVA7 A0A293M314 A0A1Z5L5L3 C1BSC9
A0A3S2LJJ0 I4DKY2 S4PAJ4 A0A0N1I5L7 A0A212FPU6 I4DK59 A0A2P8ZAF1 C4PLE9 A0A1W4XI10 Q7Q2F1 A0A2J7R6M8 Q1HQX4 A0A023EJA5 A0A023ELM8 A0A182RT68 A0A084VQF3 A0A182M718 A0A182VRF2 W5JX87 T1E2D4 A0A2M4A6F9 B3LVB0 B4GE47 A0A2M3Z171 U5EYG5 A0A1Q3EUC2 A0A1Q3EUB7 A0A1Y1KLI0 A0A2M3Z1C6 A0A182FJN9 B4PUZ3 B3P6T8 A0A1W4V8F2 A0A3B0JQK5 A0A2M4BZZ3 T1DL52 Q8SXY6 A0AQ71 A0A182IUG3 A0A1S4EL48 A0A1Q3EUD8 A0A1Q3EUC9 B4K4G5 A0A1A9W6N6 A0A336MYX4 E0VJ28 B4NIY1 B4MGF8 A0A1I8NM88 A0A1I8M6I8 A0A1A9XN09 A0A232EHB8 K7IUJ7 A0A1L8EF07 A0A1B0A8S4 A0A1B0CSV2 A0A1B0DFB6 A0A1L8DUJ1 B4JYU5 R4WQW2 A0A151WJ83 A0A195BG79 A0A195EFI6 A0A195F0Q7 F4WG56 E9IBD7 A0A195C8M4 D7GXL6 A0A1B6D7M5 A0A026WR71 A0A158NT36 A0A1B6MAZ9 A0A1J1I428 A0A0P4W5G7 E2A849 V5GN59 E2BA16 A0A3R7M9P9 A0A0J7LA60 R4UND3 A0A3L8DFM6 E9G6B6 A0A162PFE3 A0A2I9LPT8 A0A1S3II12 A0A2P2HX57 A0A1W7RAJ4 A0A0K8R831 B7P427 A0A1D2N171 C4WVA7 A0A293M314 A0A1Z5L5L3 C1BSC9
Pubmed
19121390
28756777
22651552
26354079
23622113
22118469
+ More
29403074 19405973 12364791 14747013 17210077 17204158 24945155 26483478 24438588 20920257 23761445 24330624 17994087 28004739 16951084 19126864 10731132 12537572 12537569 15327786 20566863 25315136 28648823 20075255 23691247 21719571 21282665 18362917 19820115 24508170 21347285 20798317 30249741 21292972 29248469 27289101 28528879
29403074 19405973 12364791 14747013 17210077 17204158 24945155 26483478 24438588 20920257 23761445 24330624 17994087 28004739 16951084 19126864 10731132 12537572 12537569 15327786 20566863 25315136 28648823 20075255 23691247 21719571 21282665 18362917 19820115 24508170 21347285 20798317 30249741 21292972 29248469 27289101 28528879
EMBL
BABH01039878
BABH01039879
BABH01039880
DQ443143
ABF51232.1
KZ150138
+ More
PZC72966.1 NWSH01000346 PCG77223.1 ODYU01011984 SOQ58038.1 MG846924 AXY94776.1 GDQN01011444 JAT79610.1 RSAL01000001 RVE55257.1 AK401950 KQ459324 BAM18572.1 KPJ01520.1 GAIX01004981 JAA87579.1 KQ460973 KPJ10087.1 AGBW02000323 OWR55791.1 AK401677 BAM18299.1 PYGN01000125 PSN53479.1 FM253350 CAR94540.1 AAAB01008975 EAA13499.4 NEVH01006756 PNF36492.1 DQ440320 ABF18353.1 GAPW01004552 JAC09046.1 JXUM01017578 GAPW01004434 KQ560487 JAC09164.1 KXJ82167.1 ATLV01015196 KE525003 KFB40197.1 AXCM01003940 ADMH02000068 ETN67919.1 GALA01000824 JAA94028.1 GGFK01002887 MBW36208.1 CH902617 EDV43642.1 CH479182 EDW33882.1 GGFM01001518 MBW22269.1 GANO01002014 JAB57857.1 GFDL01016134 JAV18911.1 GFDL01016135 JAV18910.1 GEZM01080398 JAV62174.1 GGFM01001497 MBW22248.1 CM000160 EDW98842.1 CH954182 EDV53758.1 OUUW01000007 SPP83253.1 GGFJ01009472 MBW58613.1 GAMD01000611 JAB00980.1 AM294798 AM294799 AM294800 AM294801 AM294802 AM294803 AM294804 AM294805 AM294806 AM294807 AM294808 FM245923 FM245924 FM245925 FM245926 FM245927 FM245928 FM245929 FM245930 FM245931 FM245932 FM245933 AE014297 AY075503 AAF56382.2 AAL68312.1 CAL26784.1 CAL26785.1 CAL26786.1 CAL26787.1 CAL26788.1 CAL26789.1 CAL26790.1 CAL26791.1 CAL26792.1 CAL26793.1 CAL26794.1 CAR93849.1 CAR93850.1 CAR93851.1 CAR93852.1 CAR93853.1 CAR93854.1 CAR93855.1 CAR93856.1 CAR93857.1 CAR93858.1 CAR93859.1 AM294809 CM000364 CAL26795.1 AXCP01008885 AXCP01008886 GFDL01016133 JAV18912.1 GFDL01016132 JAV18913.1 CH933806 EDW13917.1 UFQT01000461 UFQT01003326 SSX24584.1 SSX34875.1 DS235219 EEB13384.1 CH964272 EDW84883.1 CH940674 EDW63102.1 NNAY01004536 OXU17745.1 AAZX01002625 GFDG01001545 JAV17254.1 AJWK01026647 AJWK01026648 AJVK01015112 GFDF01003972 JAV10112.1 CH916378 EDV98560.1 AK417037 BAN20252.1 KQ983039 KYQ47929.1 KQ976488 KYM83620.1 KQ978957 KYN27008.1 KQ981897 KYN33694.1 GL888128 EGI66823.1 GL762111 EFZ22136.1 KQ978205 KYM96473.1 GG694257 EFA13494.1 GEDC01015642 JAS21656.1 KK107132 EZA58151.1 ADTU01025488 GEBQ01006875 JAT33102.1 CVRI01000039 CRK94506.1 GDRN01070703 GDRN01070698 JAI63825.1 GL437496 EFN70394.1 GALX01005469 JAB62997.1 GL446664 EFN87425.1 QCYY01001726 ROT75823.1 LBMM01000110 KMR04910.1 KC740841 AGM32665.1 QOIP01000009 RLU18698.1 GL732533 EFX85020.1 LRGB01000512 KZS18800.1 GFWZ01000427 MBW20417.1 IACF01000610 LAB66375.1 GFAH01000211 JAV48178.1 GADI01006486 JAA67322.1 ABJB010917544 DS632925 EEC01349.1 LJIJ01000321 ODM98804.1 ABLF02019504 ABLF02019508 AK341432 BAH71827.1 GFWV01009689 MAA34418.1 GFJQ02004308 JAW02662.1 BT077508 BT120851 BT121306 ACO11932.1 ADD24491.1 ADD38236.1
PZC72966.1 NWSH01000346 PCG77223.1 ODYU01011984 SOQ58038.1 MG846924 AXY94776.1 GDQN01011444 JAT79610.1 RSAL01000001 RVE55257.1 AK401950 KQ459324 BAM18572.1 KPJ01520.1 GAIX01004981 JAA87579.1 KQ460973 KPJ10087.1 AGBW02000323 OWR55791.1 AK401677 BAM18299.1 PYGN01000125 PSN53479.1 FM253350 CAR94540.1 AAAB01008975 EAA13499.4 NEVH01006756 PNF36492.1 DQ440320 ABF18353.1 GAPW01004552 JAC09046.1 JXUM01017578 GAPW01004434 KQ560487 JAC09164.1 KXJ82167.1 ATLV01015196 KE525003 KFB40197.1 AXCM01003940 ADMH02000068 ETN67919.1 GALA01000824 JAA94028.1 GGFK01002887 MBW36208.1 CH902617 EDV43642.1 CH479182 EDW33882.1 GGFM01001518 MBW22269.1 GANO01002014 JAB57857.1 GFDL01016134 JAV18911.1 GFDL01016135 JAV18910.1 GEZM01080398 JAV62174.1 GGFM01001497 MBW22248.1 CM000160 EDW98842.1 CH954182 EDV53758.1 OUUW01000007 SPP83253.1 GGFJ01009472 MBW58613.1 GAMD01000611 JAB00980.1 AM294798 AM294799 AM294800 AM294801 AM294802 AM294803 AM294804 AM294805 AM294806 AM294807 AM294808 FM245923 FM245924 FM245925 FM245926 FM245927 FM245928 FM245929 FM245930 FM245931 FM245932 FM245933 AE014297 AY075503 AAF56382.2 AAL68312.1 CAL26784.1 CAL26785.1 CAL26786.1 CAL26787.1 CAL26788.1 CAL26789.1 CAL26790.1 CAL26791.1 CAL26792.1 CAL26793.1 CAL26794.1 CAR93849.1 CAR93850.1 CAR93851.1 CAR93852.1 CAR93853.1 CAR93854.1 CAR93855.1 CAR93856.1 CAR93857.1 CAR93858.1 CAR93859.1 AM294809 CM000364 CAL26795.1 AXCP01008885 AXCP01008886 GFDL01016133 JAV18912.1 GFDL01016132 JAV18913.1 CH933806 EDW13917.1 UFQT01000461 UFQT01003326 SSX24584.1 SSX34875.1 DS235219 EEB13384.1 CH964272 EDW84883.1 CH940674 EDW63102.1 NNAY01004536 OXU17745.1 AAZX01002625 GFDG01001545 JAV17254.1 AJWK01026647 AJWK01026648 AJVK01015112 GFDF01003972 JAV10112.1 CH916378 EDV98560.1 AK417037 BAN20252.1 KQ983039 KYQ47929.1 KQ976488 KYM83620.1 KQ978957 KYN27008.1 KQ981897 KYN33694.1 GL888128 EGI66823.1 GL762111 EFZ22136.1 KQ978205 KYM96473.1 GG694257 EFA13494.1 GEDC01015642 JAS21656.1 KK107132 EZA58151.1 ADTU01025488 GEBQ01006875 JAT33102.1 CVRI01000039 CRK94506.1 GDRN01070703 GDRN01070698 JAI63825.1 GL437496 EFN70394.1 GALX01005469 JAB62997.1 GL446664 EFN87425.1 QCYY01001726 ROT75823.1 LBMM01000110 KMR04910.1 KC740841 AGM32665.1 QOIP01000009 RLU18698.1 GL732533 EFX85020.1 LRGB01000512 KZS18800.1 GFWZ01000427 MBW20417.1 IACF01000610 LAB66375.1 GFAH01000211 JAV48178.1 GADI01006486 JAA67322.1 ABJB010917544 DS632925 EEC01349.1 LJIJ01000321 ODM98804.1 ABLF02019504 ABLF02019508 AK341432 BAH71827.1 GFWV01009689 MAA34418.1 GFJQ02004308 JAW02662.1 BT077508 BT120851 BT121306 ACO11932.1 ADD24491.1 ADD38236.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000245037 UP000192223 UP000007062 UP000235965 UP000069940 UP000249989 UP000075900 UP000030765 UP000075883 UP000075920 UP000000673 UP000007801 UP000008744 UP000069272 UP000002282 UP000008711 UP000192221 UP000268350 UP000000803 UP000000304 UP000075880 UP000079169 UP000009192 UP000091820 UP000009046 UP000007798 UP000008792 UP000095300 UP000095301 UP000092443 UP000215335 UP000002358 UP000092445 UP000092461 UP000092462 UP000001070 UP000075809 UP000078540 UP000078492 UP000078541 UP000007755 UP000078542 UP000007266 UP000053097 UP000005205 UP000183832 UP000000311 UP000008237 UP000283509 UP000036403 UP000279307 UP000000305 UP000076858 UP000085678 UP000001555 UP000094527 UP000007819
UP000245037 UP000192223 UP000007062 UP000235965 UP000069940 UP000249989 UP000075900 UP000030765 UP000075883 UP000075920 UP000000673 UP000007801 UP000008744 UP000069272 UP000002282 UP000008711 UP000192221 UP000268350 UP000000803 UP000000304 UP000075880 UP000079169 UP000009192 UP000091820 UP000009046 UP000007798 UP000008792 UP000095300 UP000095301 UP000092443 UP000215335 UP000002358 UP000092445 UP000092461 UP000092462 UP000001070 UP000075809 UP000078540 UP000078492 UP000078541 UP000007755 UP000078542 UP000007266 UP000053097 UP000005205 UP000183832 UP000000311 UP000008237 UP000283509 UP000036403 UP000279307 UP000000305 UP000076858 UP000085678 UP000001555 UP000094527 UP000007819
Pfam
PF01105 EMP24_GP25L
ProteinModelPortal
Q1HQA9
A0A2W1BJG3
A0A2A4K0C5
A0A2H1WYB3
A0A3G1T1A6
A0A1E1VY15
+ More
A0A3S2LJJ0 I4DKY2 S4PAJ4 A0A0N1I5L7 A0A212FPU6 I4DK59 A0A2P8ZAF1 C4PLE9 A0A1W4XI10 Q7Q2F1 A0A2J7R6M8 Q1HQX4 A0A023EJA5 A0A023ELM8 A0A182RT68 A0A084VQF3 A0A182M718 A0A182VRF2 W5JX87 T1E2D4 A0A2M4A6F9 B3LVB0 B4GE47 A0A2M3Z171 U5EYG5 A0A1Q3EUC2 A0A1Q3EUB7 A0A1Y1KLI0 A0A2M3Z1C6 A0A182FJN9 B4PUZ3 B3P6T8 A0A1W4V8F2 A0A3B0JQK5 A0A2M4BZZ3 T1DL52 Q8SXY6 A0AQ71 A0A182IUG3 A0A1S4EL48 A0A1Q3EUD8 A0A1Q3EUC9 B4K4G5 A0A1A9W6N6 A0A336MYX4 E0VJ28 B4NIY1 B4MGF8 A0A1I8NM88 A0A1I8M6I8 A0A1A9XN09 A0A232EHB8 K7IUJ7 A0A1L8EF07 A0A1B0A8S4 A0A1B0CSV2 A0A1B0DFB6 A0A1L8DUJ1 B4JYU5 R4WQW2 A0A151WJ83 A0A195BG79 A0A195EFI6 A0A195F0Q7 F4WG56 E9IBD7 A0A195C8M4 D7GXL6 A0A1B6D7M5 A0A026WR71 A0A158NT36 A0A1B6MAZ9 A0A1J1I428 A0A0P4W5G7 E2A849 V5GN59 E2BA16 A0A3R7M9P9 A0A0J7LA60 R4UND3 A0A3L8DFM6 E9G6B6 A0A162PFE3 A0A2I9LPT8 A0A1S3II12 A0A2P2HX57 A0A1W7RAJ4 A0A0K8R831 B7P427 A0A1D2N171 C4WVA7 A0A293M314 A0A1Z5L5L3 C1BSC9
A0A3S2LJJ0 I4DKY2 S4PAJ4 A0A0N1I5L7 A0A212FPU6 I4DK59 A0A2P8ZAF1 C4PLE9 A0A1W4XI10 Q7Q2F1 A0A2J7R6M8 Q1HQX4 A0A023EJA5 A0A023ELM8 A0A182RT68 A0A084VQF3 A0A182M718 A0A182VRF2 W5JX87 T1E2D4 A0A2M4A6F9 B3LVB0 B4GE47 A0A2M3Z171 U5EYG5 A0A1Q3EUC2 A0A1Q3EUB7 A0A1Y1KLI0 A0A2M3Z1C6 A0A182FJN9 B4PUZ3 B3P6T8 A0A1W4V8F2 A0A3B0JQK5 A0A2M4BZZ3 T1DL52 Q8SXY6 A0AQ71 A0A182IUG3 A0A1S4EL48 A0A1Q3EUD8 A0A1Q3EUC9 B4K4G5 A0A1A9W6N6 A0A336MYX4 E0VJ28 B4NIY1 B4MGF8 A0A1I8NM88 A0A1I8M6I8 A0A1A9XN09 A0A232EHB8 K7IUJ7 A0A1L8EF07 A0A1B0A8S4 A0A1B0CSV2 A0A1B0DFB6 A0A1L8DUJ1 B4JYU5 R4WQW2 A0A151WJ83 A0A195BG79 A0A195EFI6 A0A195F0Q7 F4WG56 E9IBD7 A0A195C8M4 D7GXL6 A0A1B6D7M5 A0A026WR71 A0A158NT36 A0A1B6MAZ9 A0A1J1I428 A0A0P4W5G7 E2A849 V5GN59 E2BA16 A0A3R7M9P9 A0A0J7LA60 R4UND3 A0A3L8DFM6 E9G6B6 A0A162PFE3 A0A2I9LPT8 A0A1S3II12 A0A2P2HX57 A0A1W7RAJ4 A0A0K8R831 B7P427 A0A1D2N171 C4WVA7 A0A293M314 A0A1Z5L5L3 C1BSC9
PDB
5AZY
E-value=2.19464e-19,
Score=231
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
Endoplasmic reticulum membrane
Endoplasmic reticulum membrane
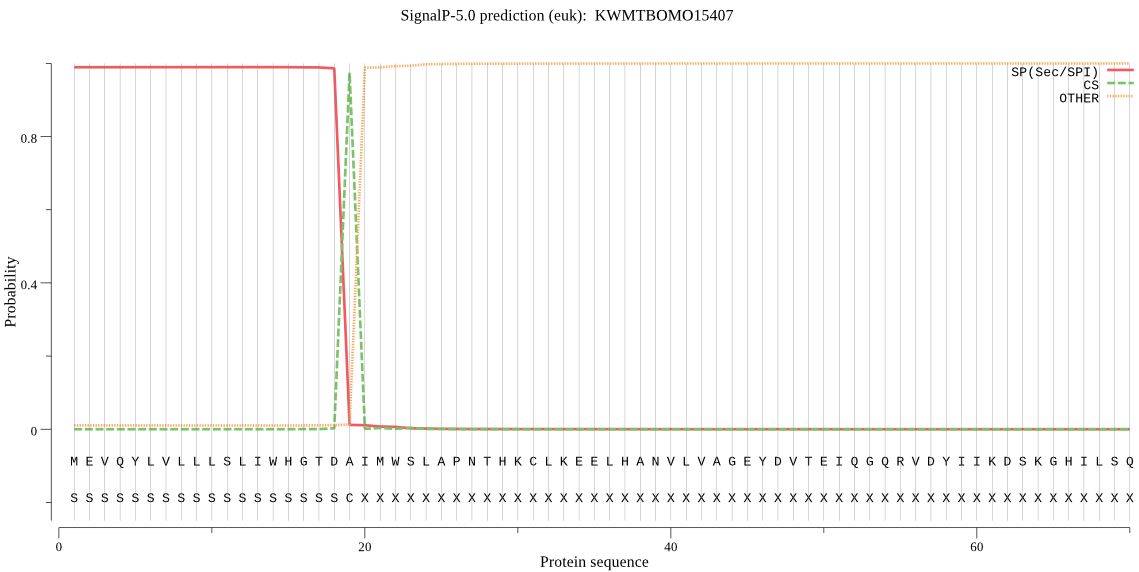
SignalP
Position: 1 - 19,
Likelihood: 0.989081
Length:
205
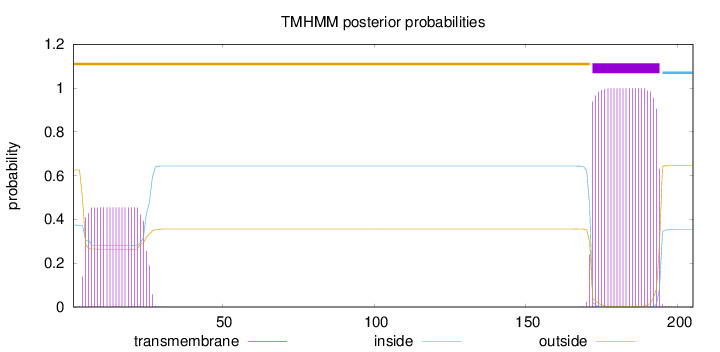
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.19301
Exp number, first 60 AAs:
9.58656999999999
Total prob of N-in:
0.37470
outside
1 - 171
TMhelix
172 - 194
inside
195 - 205
Population Genetic Test Statistics
Pi
213.384096
Theta
188.928117
Tajima's D
0.137992
CLR
153.347957
CSRT
0.403529823508825
Interpretation
Uncertain
