Gene
KWMTBOMO15405 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004892
Annotation
TIA-1_homologue_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.828
Sequence
CDS
ATGGGCGACGAAAGCCACCCGAAAACTCTCTACGTCGGTAATTTGGACCCGAGTGTCACAGAAGTTTTCCTATGTACGTTGTTTGGTCAAATAGGCGATGTAAAAGGTTGCAAGATTATACGGGAGCCAGGAAATGACCCGTATGCATTTCTCGAGTTTACGTGTCACACGGCGGCGGCCACAGCGCTGGCCGCCATGAACAAGCGAGTGGTCCTCGACAAGGAAATGAAGGTTAATTGGGCGACGAGTCCTGGTAACCAGCCGAAGACGGACACTAGTAATCATCATCACATATTCGTGGGCGATCTGTCTCCAGAAATCGAAACAAACATACTCCGAGAGGCCTTCGCTCCATTTGGTGAGATATCGAACTGTCGCATAGTACGCGATCCTCAAACGCTCAAATCCAAGGGTTATGCTTTTGTATCATTTGTAAAGAAAGCAGATGCCGAAGCTGCCATCCAGGCAATGAATGGCCAGTGGCTGGGATCAAGATCCATACGTACGAACTGGTCAACGCGTAAACCACCAGCGAAGGGTGTGAATGAAGGCGCGCCCAGTAGTAAGAGAGCAAAACAGCCAACATTTGACGAAGTTTATAATCAAAGCTCACCGACAAACACCACAGTTTACTGTGGCGGTTTCACAAGCAACATTATTACTGAAGAGTTAATGCAGAACACATTCTCACAATTTGGTCAGATACAGGACATCAGAGTTTTCAGAGATAAGGGCTATGCATTCATTAGGTTTACTACAAAGGAAGCTGCAGCACATGCCATAGAAGCTACACACAATACAGAAATAAGCGGTCACACAGTCAAATGTTTCTGGGGGAAAGAGAATGGAGGTGCTGAAAACCAGGTTCGACACACGGCAGACTCGAACAGCACGGTCCCAGATCAGAGCGGGAACAACTCGTCGGCGGCGCCTCCAGCGATGGGCGGCCAGTCCCAATATCCTTACCCGTATCAGCAAGGAATGGGCTATTGGTATGCTCAGGGTTACCCTGCGTTACAGGGCTACATGGCTCCCGGGTACTACCAGCAGTACGCGGCCGCGTACAGCAACCCTCAGGCCGCCGCGGCAGCGGGGTACCGCATGGGAATGGGCGGTAATGGCGGCGGCGGCGCCGGGTGGGGCGGCCCCTCGCAACCCCTCATGTACGCGTCGGCCGCCATGCCCTCCGCGCAGTACCCCTCCCAGTAG
Protein
MGDESHPKTLYVGNLDPSVTEVFLCTLFGQIGDVKGCKIIREPGNDPYAFLEFTCHTAAATALAAMNKRVVLDKEMKVNWATSPGNQPKTDTSNHHHIFVGDLSPEIETNILREAFAPFGEISNCRIVRDPQTLKSKGYAFVSFVKKADAEAAIQAMNGQWLGSRSIRTNWSTRKPPAKGVNEGAPSSKRAKQPTFDEVYNQSSPTNTTVYCGGFTSNIITEELMQNTFSQFGQIQDIRVFRDKGYAFIRFTTKEAAAHAIEATHNTEISGHTVKCFWGKENGGAENQVRHTADSNSTVPDQSGNNSSAAPPAMGGQSQYPYPYQQGMGYWYAQGYPALQGYMAPGYYQQYAAAYSNPQAAAAAGYRMGMGGNGGGGAGWGGPSQPLMYASAAMPSAQYPSQ
Summary
Uniprot
C8KHM3
C8KHM2
Q0N2S1
H9J5V2
Q76KD3
A0A2W1BH06
+ More
A0A194Q7M0 A0A2H1WI71 C0SQ69 A0A3S2P282 A0A2A4JYY1 A0A212FQ18 A0A0N0PB93 C5NMH3 A0A0L7LP39 D6WSZ4 V5I9A6 A0A1Y1KBA5 S4NJ30 A0A0T6AXQ7 J3JYC2 A0A2P8XNW9 A0A1B0GMD7 A0A1L8DH14 A0A1L8DHD0 A0A1B6D785 A0A2J7Q259 A0A1B0CMT7 A0A1B6IIL9 A0A1B6LRL7 A0A1B6GDF4 A0A067QTH3 A0A224XC23 A0A026WSF1 A0A195DVL6 E2AI23 F4W8A5 A0A151I5Z6 A0A195EWU1 A0A158NSN7 A0A151XDV7 A0A224XDP3 A0A310SJP0 A0A084WA20 A0A195CD20 W5JUE3 A0A2M3Z227 A0A182IKZ8 A0A2M4ACE4 A0A0L7R701 A0A182F6J4 A0A2M4BMI5 A0A0J7KPR5 A0A1S4ER17 A0A2A3E6C4 A0A088A059 A0A0N0BE34 A0A182QN73 A0A2A3E5P1 A0A034WU24 A0A232F552 T1HFG1 A0A1Q3F0M3 A0A1B0BJP5 B0X286 A0A1Q3F0K3 U5EX85 A0A0K8V269 W8AS17 W8AGB4 A0A0K8UHD0 A0A0A1XNF5 A0A0A1XLH7 T1PAQ1 A0A1I8MD80 A0A182TIU1 A0A182XEI8 A0A182HTD4 Q175B9 A0A154P4X1 E0VYI5 A0A182LFI5 Q7QBW1 A0A1B0G863 A0A240SX76 A0A0L0CJB3 A0A1A9X1B3 A0A146LK12 A0A182P868 A0A182KH25 A0A1I8P6I4 A0A2H8TUQ6 A0A2S2PKR1 A0A0R3NGZ0
A0A194Q7M0 A0A2H1WI71 C0SQ69 A0A3S2P282 A0A2A4JYY1 A0A212FQ18 A0A0N0PB93 C5NMH3 A0A0L7LP39 D6WSZ4 V5I9A6 A0A1Y1KBA5 S4NJ30 A0A0T6AXQ7 J3JYC2 A0A2P8XNW9 A0A1B0GMD7 A0A1L8DH14 A0A1L8DHD0 A0A1B6D785 A0A2J7Q259 A0A1B0CMT7 A0A1B6IIL9 A0A1B6LRL7 A0A1B6GDF4 A0A067QTH3 A0A224XC23 A0A026WSF1 A0A195DVL6 E2AI23 F4W8A5 A0A151I5Z6 A0A195EWU1 A0A158NSN7 A0A151XDV7 A0A224XDP3 A0A310SJP0 A0A084WA20 A0A195CD20 W5JUE3 A0A2M3Z227 A0A182IKZ8 A0A2M4ACE4 A0A0L7R701 A0A182F6J4 A0A2M4BMI5 A0A0J7KPR5 A0A1S4ER17 A0A2A3E6C4 A0A088A059 A0A0N0BE34 A0A182QN73 A0A2A3E5P1 A0A034WU24 A0A232F552 T1HFG1 A0A1Q3F0M3 A0A1B0BJP5 B0X286 A0A1Q3F0K3 U5EX85 A0A0K8V269 W8AS17 W8AGB4 A0A0K8UHD0 A0A0A1XNF5 A0A0A1XLH7 T1PAQ1 A0A1I8MD80 A0A182TIU1 A0A182XEI8 A0A182HTD4 Q175B9 A0A154P4X1 E0VYI5 A0A182LFI5 Q7QBW1 A0A1B0G863 A0A240SX76 A0A0L0CJB3 A0A1A9X1B3 A0A146LK12 A0A182P868 A0A182KH25 A0A1I8P6I4 A0A2H8TUQ6 A0A2S2PKR1 A0A0R3NGZ0
Pubmed
19121390
14597390
28756777
26354079
19270389
22118469
+ More
26227816 18362917 19820115 28004739 23622113 22516182 23537049 29403074 24845553 24508170 30249741 20798317 21719571 21347285 24438588 20920257 23761445 25348373 28648823 24495485 25830018 25315136 17510324 20566863 20966253 12364791 14747013 17210077 26108605 26823975 15632085 17994087
26227816 18362917 19820115 28004739 23622113 22516182 23537049 29403074 24845553 24508170 30249741 20798317 21719571 21347285 24438588 20920257 23761445 25348373 28648823 24495485 25830018 25315136 17510324 20566863 20966253 12364791 14747013 17210077 26108605 26823975 15632085 17994087
EMBL
AB461842
BAI40366.1
AB461841
BAI40365.1
DQ646410
ABH10800.1
+ More
BABH01039881 BABH01039882 AB092510 BAD00701.1 KZ150138 PZC72964.1 KQ459324 KPJ01518.1 ODYU01008821 SOQ52759.1 AB331732 BAH56564.1 RSAL01000001 RVE55255.1 NWSH01000346 PCG77225.1 AGBW02000323 OWR55789.1 KQ460973 KPJ10085.1 AB331733 BAH84828.1 JTDY01000450 KOB77124.1 KQ971352 EFA06678.1 GALX01003270 JAB65196.1 GEZM01087307 JAV58783.1 GAIX01013844 JAA78716.1 LJIG01022612 KRT79637.1 APGK01055582 BT128248 KB741266 KB632194 AEE63208.1 ENN71538.1 ERL89908.1 PYGN01001626 PSN33700.1 AJVK01024436 AJVK01024437 GFDF01008335 JAV05749.1 GFDF01008299 JAV05785.1 GEDC01015754 JAS21544.1 NEVH01019373 PNF22673.1 AJWK01019157 AJWK01019158 AJWK01019159 AJWK01019160 AJWK01019161 AJWK01019162 GECU01020938 JAS86768.1 GEBQ01013607 JAT26370.1 GECZ01009287 JAS60482.1 KK853291 KDR08866.1 GFTR01006460 JAW09966.1 KK107111 QOIP01000011 EZA58903.1 RLU16972.1 KQ980322 KYN16604.1 GL439621 EFN66931.1 GL887898 EGI69600.1 KQ976400 KYM92669.1 KQ981948 KYN32616.1 ADTU01025080 KQ982275 KYQ58438.1 GFTR01005869 JAW10557.1 KQ764717 OAD54416.1 ATLV01022002 KE525327 KFB47064.1 KQ978023 KYM97978.1 ADMH02000060 ETN67962.1 GGFM01001819 MBW22570.1 GGFK01005132 MBW38453.1 KQ414643 KOC66662.1 GGFJ01005101 MBW54242.1 LBMM01004593 KMQ92261.1 KZ288379 PBC26601.1 KQ435848 KOX70991.1 AXCN02001613 PBC26602.1 GAKP01001111 GAKP01001110 JAC57842.1 NNAY01000955 OXU25742.1 ACPB03024322 GFDL01014007 JAV21038.1 JXJN01015475 DS232283 EDS39109.1 GFDL01014027 JAV21018.1 GANO01000190 JAB59681.1 GDHF01019441 JAI32873.1 GAMC01019182 GAMC01019181 JAB87373.1 GAMC01019183 GAMC01019180 JAB87375.1 GDHF01026998 GDHF01026240 GDHF01025837 GDHF01008509 GDHF01006542 JAI25316.1 JAI26074.1 JAI26477.1 JAI43805.1 JAI45772.1 GBXI01008952 GBXI01002179 JAD05340.1 JAD12113.1 GBXI01002864 JAD11428.1 KA645817 AFP60446.1 CH477403 EAT41668.1 KQ434809 KZC06364.1 DS235845 EEB18441.1 AAAB01008859 EAA07505.5 CCAG010000199 JRES01000321 KNC32307.1 GDHC01010195 JAQ08434.1 GFXV01005736 MBW17541.1 GGMR01017432 MBY30051.1 CM000070 KRT00354.1
BABH01039881 BABH01039882 AB092510 BAD00701.1 KZ150138 PZC72964.1 KQ459324 KPJ01518.1 ODYU01008821 SOQ52759.1 AB331732 BAH56564.1 RSAL01000001 RVE55255.1 NWSH01000346 PCG77225.1 AGBW02000323 OWR55789.1 KQ460973 KPJ10085.1 AB331733 BAH84828.1 JTDY01000450 KOB77124.1 KQ971352 EFA06678.1 GALX01003270 JAB65196.1 GEZM01087307 JAV58783.1 GAIX01013844 JAA78716.1 LJIG01022612 KRT79637.1 APGK01055582 BT128248 KB741266 KB632194 AEE63208.1 ENN71538.1 ERL89908.1 PYGN01001626 PSN33700.1 AJVK01024436 AJVK01024437 GFDF01008335 JAV05749.1 GFDF01008299 JAV05785.1 GEDC01015754 JAS21544.1 NEVH01019373 PNF22673.1 AJWK01019157 AJWK01019158 AJWK01019159 AJWK01019160 AJWK01019161 AJWK01019162 GECU01020938 JAS86768.1 GEBQ01013607 JAT26370.1 GECZ01009287 JAS60482.1 KK853291 KDR08866.1 GFTR01006460 JAW09966.1 KK107111 QOIP01000011 EZA58903.1 RLU16972.1 KQ980322 KYN16604.1 GL439621 EFN66931.1 GL887898 EGI69600.1 KQ976400 KYM92669.1 KQ981948 KYN32616.1 ADTU01025080 KQ982275 KYQ58438.1 GFTR01005869 JAW10557.1 KQ764717 OAD54416.1 ATLV01022002 KE525327 KFB47064.1 KQ978023 KYM97978.1 ADMH02000060 ETN67962.1 GGFM01001819 MBW22570.1 GGFK01005132 MBW38453.1 KQ414643 KOC66662.1 GGFJ01005101 MBW54242.1 LBMM01004593 KMQ92261.1 KZ288379 PBC26601.1 KQ435848 KOX70991.1 AXCN02001613 PBC26602.1 GAKP01001111 GAKP01001110 JAC57842.1 NNAY01000955 OXU25742.1 ACPB03024322 GFDL01014007 JAV21038.1 JXJN01015475 DS232283 EDS39109.1 GFDL01014027 JAV21018.1 GANO01000190 JAB59681.1 GDHF01019441 JAI32873.1 GAMC01019182 GAMC01019181 JAB87373.1 GAMC01019183 GAMC01019180 JAB87375.1 GDHF01026998 GDHF01026240 GDHF01025837 GDHF01008509 GDHF01006542 JAI25316.1 JAI26074.1 JAI26477.1 JAI43805.1 JAI45772.1 GBXI01008952 GBXI01002179 JAD05340.1 JAD12113.1 GBXI01002864 JAD11428.1 KA645817 AFP60446.1 CH477403 EAT41668.1 KQ434809 KZC06364.1 DS235845 EEB18441.1 AAAB01008859 EAA07505.5 CCAG010000199 JRES01000321 KNC32307.1 GDHC01010195 JAQ08434.1 GFXV01005736 MBW17541.1 GGMR01017432 MBY30051.1 CM000070 KRT00354.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000007151
UP000053240
+ More
UP000037510 UP000007266 UP000019118 UP000030742 UP000245037 UP000092462 UP000235965 UP000092461 UP000027135 UP000053097 UP000279307 UP000078492 UP000000311 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000030765 UP000078542 UP000000673 UP000075880 UP000053825 UP000069272 UP000036403 UP000079169 UP000242457 UP000005203 UP000053105 UP000075886 UP000215335 UP000015103 UP000092460 UP000002320 UP000095301 UP000075902 UP000076407 UP000008820 UP000076502 UP000009046 UP000075882 UP000007062 UP000092444 UP000092445 UP000037069 UP000091820 UP000075885 UP000075881 UP000095300 UP000001819
UP000037510 UP000007266 UP000019118 UP000030742 UP000245037 UP000092462 UP000235965 UP000092461 UP000027135 UP000053097 UP000279307 UP000078492 UP000000311 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000030765 UP000078542 UP000000673 UP000075880 UP000053825 UP000069272 UP000036403 UP000079169 UP000242457 UP000005203 UP000053105 UP000075886 UP000215335 UP000015103 UP000092460 UP000002320 UP000095301 UP000075902 UP000076407 UP000008820 UP000076502 UP000009046 UP000075882 UP000007062 UP000092444 UP000092445 UP000037069 UP000091820 UP000075885 UP000075881 UP000095300 UP000001819
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
C8KHM3
C8KHM2
Q0N2S1
H9J5V2
Q76KD3
A0A2W1BH06
+ More
A0A194Q7M0 A0A2H1WI71 C0SQ69 A0A3S2P282 A0A2A4JYY1 A0A212FQ18 A0A0N0PB93 C5NMH3 A0A0L7LP39 D6WSZ4 V5I9A6 A0A1Y1KBA5 S4NJ30 A0A0T6AXQ7 J3JYC2 A0A2P8XNW9 A0A1B0GMD7 A0A1L8DH14 A0A1L8DHD0 A0A1B6D785 A0A2J7Q259 A0A1B0CMT7 A0A1B6IIL9 A0A1B6LRL7 A0A1B6GDF4 A0A067QTH3 A0A224XC23 A0A026WSF1 A0A195DVL6 E2AI23 F4W8A5 A0A151I5Z6 A0A195EWU1 A0A158NSN7 A0A151XDV7 A0A224XDP3 A0A310SJP0 A0A084WA20 A0A195CD20 W5JUE3 A0A2M3Z227 A0A182IKZ8 A0A2M4ACE4 A0A0L7R701 A0A182F6J4 A0A2M4BMI5 A0A0J7KPR5 A0A1S4ER17 A0A2A3E6C4 A0A088A059 A0A0N0BE34 A0A182QN73 A0A2A3E5P1 A0A034WU24 A0A232F552 T1HFG1 A0A1Q3F0M3 A0A1B0BJP5 B0X286 A0A1Q3F0K3 U5EX85 A0A0K8V269 W8AS17 W8AGB4 A0A0K8UHD0 A0A0A1XNF5 A0A0A1XLH7 T1PAQ1 A0A1I8MD80 A0A182TIU1 A0A182XEI8 A0A182HTD4 Q175B9 A0A154P4X1 E0VYI5 A0A182LFI5 Q7QBW1 A0A1B0G863 A0A240SX76 A0A0L0CJB3 A0A1A9X1B3 A0A146LK12 A0A182P868 A0A182KH25 A0A1I8P6I4 A0A2H8TUQ6 A0A2S2PKR1 A0A0R3NGZ0
A0A194Q7M0 A0A2H1WI71 C0SQ69 A0A3S2P282 A0A2A4JYY1 A0A212FQ18 A0A0N0PB93 C5NMH3 A0A0L7LP39 D6WSZ4 V5I9A6 A0A1Y1KBA5 S4NJ30 A0A0T6AXQ7 J3JYC2 A0A2P8XNW9 A0A1B0GMD7 A0A1L8DH14 A0A1L8DHD0 A0A1B6D785 A0A2J7Q259 A0A1B0CMT7 A0A1B6IIL9 A0A1B6LRL7 A0A1B6GDF4 A0A067QTH3 A0A224XC23 A0A026WSF1 A0A195DVL6 E2AI23 F4W8A5 A0A151I5Z6 A0A195EWU1 A0A158NSN7 A0A151XDV7 A0A224XDP3 A0A310SJP0 A0A084WA20 A0A195CD20 W5JUE3 A0A2M3Z227 A0A182IKZ8 A0A2M4ACE4 A0A0L7R701 A0A182F6J4 A0A2M4BMI5 A0A0J7KPR5 A0A1S4ER17 A0A2A3E6C4 A0A088A059 A0A0N0BE34 A0A182QN73 A0A2A3E5P1 A0A034WU24 A0A232F552 T1HFG1 A0A1Q3F0M3 A0A1B0BJP5 B0X286 A0A1Q3F0K3 U5EX85 A0A0K8V269 W8AS17 W8AGB4 A0A0K8UHD0 A0A0A1XNF5 A0A0A1XLH7 T1PAQ1 A0A1I8MD80 A0A182TIU1 A0A182XEI8 A0A182HTD4 Q175B9 A0A154P4X1 E0VYI5 A0A182LFI5 Q7QBW1 A0A1B0G863 A0A240SX76 A0A0L0CJB3 A0A1A9X1B3 A0A146LK12 A0A182P868 A0A182KH25 A0A1I8P6I4 A0A2H8TUQ6 A0A2S2PKR1 A0A0R3NGZ0
PDB
2MJN
E-value=3.95327e-61,
Score=595
Ontologies
GO
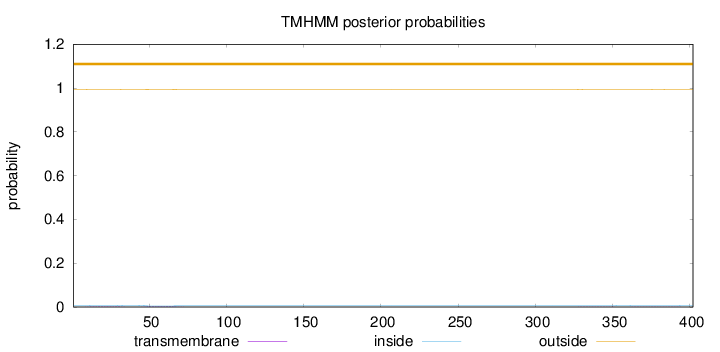
Topology
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11269
Exp number, first 60 AAs:
0.07323
Total prob of N-in:
0.00700
outside
1 - 402
Population Genetic Test Statistics
Pi
169.328056
Theta
170.948094
Tajima's D
-0.596806
CLR
2.386787
CSRT
0.218889055547223
Interpretation
Possibly Positive selection
